













| General Information: |
|
| Name(s) found: |
MRE11_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 720 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
DNA metabolic process
[IEA]
double-strand break repair [IEA] |
| Molecular Function: |
protein serine/threonine phosphatase activity
[ISS]
exonuclease activity [IEA] endonuclease activity [IEA] manganese ion binding [IEA] hydrolase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSREDFSDTL RVLVATDCHL GYMEKDEIRR HDSFKAFEEI CSIAEEKQVD FLLLGGDLFH 60
61 ENKPSRTTLV KAIEILRRHC LNDKPVQFQV VSDQTVNFQN AFGQVNYEDP HFNVGLPVFS 120
121 IHGNHDDPAG VDNLSAIDIL SACNLVNYFG KMVLGGSGVG QITLYPILMK KGSTTVALYG 180
181 LGNIRDERLN RMFQTPHAVQ WMRPEVQEGC DVSDWFNILV LHQNRVKSNP KNAISEHFLP 240
241 RFLDFIVWGH EHECLIDPQE VSGMGFHITQ PGSSVATSLI DGESKPKHVL LLEIKGNQYR 300
301 PTKIPLTSVR PFEYTEIVLK DESDIDPNDQ NSILEHLDKV VRNLIEKASK KAVNRSEIKL 360
361 PLVRIKVDYS GFMTINPQRF GQKYVGKVAN PQDILIFSKA SKKGRSEANI DDSERLRPEE 420
421 LNQQNIEALV AESNLKMEIL PVNDLDVALH NFVNKDDKLA FYSCVQYNLQ ETRGKLAKDS 480
481 DAKKFEEDDL ILKVGECLEE RLKDRSTRPT GSSQFLSTGL TSENLTKGSS GIANASFSDD 540
541 EDTTQMSGLA PPTRGRRGSS TANTTRGRAK APTRGRGRGK ASSAMKQTTL DSSLGFRQSQ 600
601 RSASAAASAA FKSASTIGED DVDSPSSEEV EPEDFNKPDS SSEDDESTKG KGRKRPATTK 660
661 RGRGRGSGTS KRGRKNESSS SLNRLLSSKD DDEDEDDEDR EKKLNKSQPR VTRNYGALRR 720
721 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
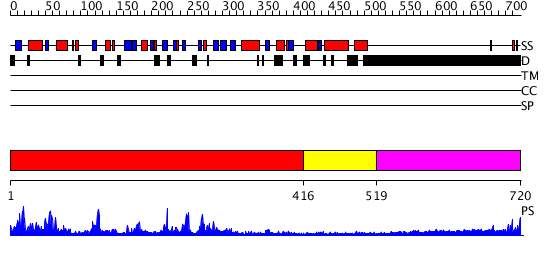
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..415] | 47.0 | Mre11 | |
| 2 | View Details | [416..518] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [519..720] | 11.199997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)