













| General Information: |
|
| Name(s) found: |
MDHG2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 354 amino acids |
Gene Ontology: |
|
| Cellular Component: |
peroxisome
[IDA]
chloroplast [IDA] |
| Biological Process: |
regulation of photorespiration
[IMP]
regulation of fatty acid beta-oxidation [IGI][IMP] |
| Molecular Function: |
L-malate dehydrogenase activity
[IEA]
malate dehydrogenase activity [IEA][ISS] oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor [IEA] oxidoreductase activity [IEA] catalytic activity [IEA] binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDPNQRIARI SAHLNPPNLH NQIADGSGLN RVACRAKGGS PGFKVAILGA AGGIGQPLAM 60
61 LMKMNPLVSV LHLYDVANAP GVTADISHMD TSAVVRGFLG QPQLEEALTG MDLVIIPAGV 120
121 PRKPGMTRDD LFNINAGIVR TLSEAIAKCC PKAIVNIISN PVNSTVPIAA EVFKKAGTFD 180
181 PKKLMGVTML DVVRANTFVA EVMSLDPREV EVPVVGGHAG VTILPLLSQV KPPCSFTQKE 240
241 IEYLTDRIQN GGTEVVEAKA GAGSATLSMA YAAVEFADAC LRGLRGDANI VECAYVASHV 300
301 TELPFFASKV RLGRCGIDEV YGLGPLNEYE RMGLEKAKKE LSVSIHKGVT FAKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
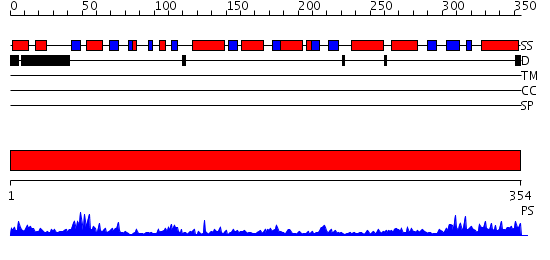
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..354] | 79.221849 | Mature and translocatable forms of glyoxysomal malate dehydrogenase have different activities and stabilities but similar crystal structures |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.67 |
Source: Reynolds et al. (2008)