













| General Information: |
|
| Name(s) found: |
LTI78_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 710 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
response to cold
[IEP]
hyperosmotic salinity response [IEP] response to abscisic acid stimulus [IEP] response to salt stress [IEP] response to osmotic stress [IGI] response to desiccation [IMP] response to water deprivation [IEP] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDQTEEPPLN THQQHPEEVE HHENGATKMF RKVKARAKKF KNSLTKHGQS NEHEQDHDLV 60
61 EEDDDDDELE PEVIDAPGVT GKPRETNVPA SEEIIPPGTK VFPVVSSDYT KPTESVPVQE 120
121 ASYGHDAPAH SVRTTFTSDK EEKRDVPIHH PLSELSDREE SRETHHESLN TPVSLLSGTE 180
181 DVTSTFAPSG DDEYLDGQRK VNVETPITLE EESAVSDYLS GVSNYQSKVT DPTKEETGGV 240
241 PEIAESFGNM EVTDESPDQK PGQFERDLST RSKEFKEFDQ DFDSVLGKDS PAKFPGESGV 300
301 VFPVGFGDES GAELEKDFPT RSHDFDMKTE TGMDTNSPSR SHEFDLKTES GNDKNSPMGF 360
361 GSESGAELEK EFDQKNDSGR NEYSPESDGG LGAPLGGNFP VRSHELDLKN ESDIDKDVPT 420
421 GFDGEPDFLA KGRPGYGEAS EEDKFPARSD DVEVETELGR DPKTETLDQF SPELSHPKER 480
481 DEFKESRDDF EETRDEKTEE PKQSTYTEKF ASMLGYSGEI PVGDQTQVAG TVDEKLTPVN 540
541 EKDQETESAV TTKLPISGGG SGVEEQRGED KSVSGRDYVA EKLTTEEEDK AFSDMVAEKL 600
601 QIGGEEEKKE TTTKEVEKIS TEKAASEEGE AVEEEVKGGG GMVGRIKGWF GGGATDEVKP 660
661 ESPHSVEEAP KSSGWFGGGA TEEVKPKSPH SVEESPQSLG STVVPVQKEL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
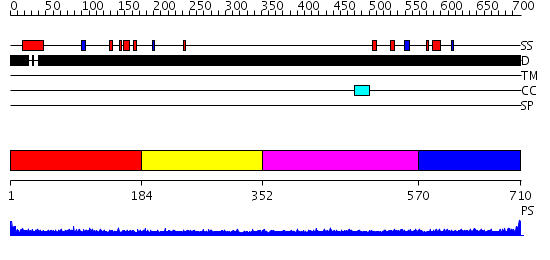
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..183] | 2.208997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [184..351] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [352..569] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [570..710] | 2.203994 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)