













| General Information: |
|
| Name(s) found: |
LTI65_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 619 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
response to cold
[IEP]
abscisic acid mediated signaling pathway [IEP] response to abscisic acid stimulus [IEP] response to salt stress [IEP] response to water deprivation [IEP] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MESQLTRPYG HEQAEEPIRI HHPEEEEHHE KGASKVLKKV KEKAKKIKNS LTKHGNGHDH 60
61 DVEDDDDEYD EQDPEVHGAP VYESSAVRGG VTGKPKSLSH AGETNVPASE EIVPPGTKVF 120
121 PVVSSDHTKP IEPVSLQDTS YGHEALADPV RTTETSDWEA KREAPTHYPL GVSEFSDRGE 180
181 SREAHQEPLN TPVSLLSATE DVTRTFAPGG EDDYLGGQRK VNVETPKRLE EDPAAPGGGS 240
241 DYLSGVSNYQ SKVTDPTHKG GEAGVPEIAE SLGRMKVTDE SPDQKSRQGR EEDFPTRSHE 300
301 FDLKKESDIN KNSPARFGGE SKAGMEEDFP TRGDVKVESG LGRDLPTGTH DQFSPELSRP 360
361 KERDDSEETK DESTHETKPS TYTEQLASAT SAITNKAIAA KNVVASKLGY TGENGGGQSE 420
421 SPVKDETPRS VTAYGQKVAG TVAEKLTPVY EKVKETGSTV MTKLPLSGGG SGVKETQQGE 480
481 EKGVTAKNYI SEKLKPGEED KALSEMIAEK LHFGGGGEKK TTATKEVEVT VEKIPSDQIA 540
541 EGKGHGEAVA EEGKGGEGMV GKVKGAVTSW LGGKPKSPRS VEESPQSLGT TVGTMGFSDS 600
601 GGSELGGSGG GKGVQDSGN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
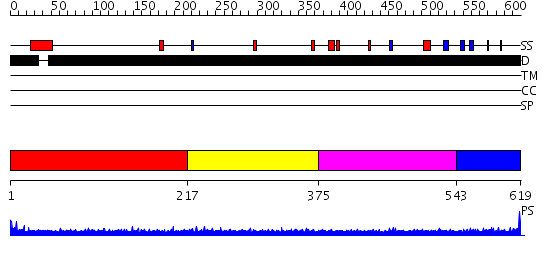
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..216] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [217..374] | 2.191995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [375..542] | 1.183996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [543..619] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)