













| General Information: |
|
| Name(s) found: |
KPRS2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 400 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IEA]
|
| Biological Process: |
ribonucleoside monophosphate biosynthetic process
[IEA]
nucleotide biosynthetic process [IEA][ISS] nucleoside metabolic process [IEA] cellular biosynthetic process [IEA] |
| Molecular Function: |
magnesium ion binding
[IEA]
ribose phosphate diphosphokinase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASLALTSPP SVKIPSYLSS SSSSLFSRSS ISFRTTESRS RICVSGYAKC NLPKALNGNA 60
61 RVPIINETTI PKFFDSSRLE KSVSRNNTKL KLFSGTANPA LSQEIAWYMG LELGKVSIKR 120
121 FADGEIYVQL KESVRGCDVF LVQPTCTPTN ENLMELLIMV DACRRASAKK VTAVIPYFGY 180
181 ARADRKTQGR ESIAAKLVAN LITEAGADRV LACDLHSGQS MGYFDIPVDH VYCQPVILDY 240
241 LASKSISSED LVVVSPDVGG VARARAFAKK LSDAPLAIVD KRRHGHNVAE VMNLIGDVKG 300
301 KVAVMVDDII DTAGTIVKGA ALLHEEGARE VYACCTHAVF SPPAIERLSS GLLQEVIVTN 360
361 TLPVAEKNYF PQLTILSVAN LLGETIWRVH DDSSVSSIFL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
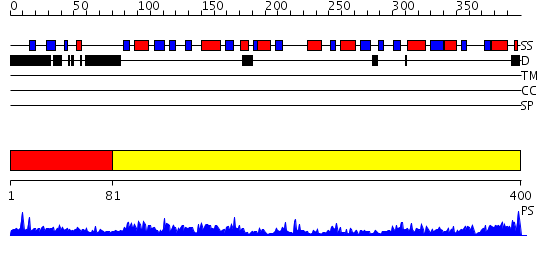
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | 1.089995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [81..400] | 104.0 | Phosphoribosylpyrophosphate synthetase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.60 |
Source: Reynolds et al. (2008)