













| General Information: |
|
| Name(s) found: |
HAT2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 283 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA][ISS]
|
| Biological Process: |
unidimensional cell growth
[IMP]
regulation of transcription, DNA-dependent [ISS] shade avoidance [IEP] auxin mediated signaling pathway [IEP] response to auxin stimulus [IMP] |
| Molecular Function: |
DNA binding
[ISS]
transcription factor activity [ISS] transcription repressor activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMMGKEDLGL SLSLGFSQNH NPLQMNLNPN SSLSNNLQRL PWNQTFDPTS DLRKIDVNSF 60
61 PSTVNCEEDT GVSSPNSTIS STISGKRSER EGISGTGVGS GDDHDEITPD RGYSRGTSDE 120
121 EEDGGETSRK KLRLSKDQSA FLEETFKEHN TLNPKQKLAL AKKLNLTARQ VEVWFQNRRA 180
181 RTKLKQTEVD CEYLKRCVEK LTEENRRLQK EAMELRTLKL SPQFYGQMTP PTTLIMCPSC 240
241 ERVGGPSSSN HHHNHRPVSI NPWVACAGQV AHGLNFEALR PRS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
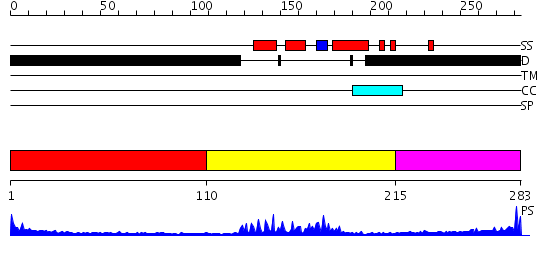
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..109] | 46.619789 | No description for PF04618.3 was found. No confident structure predictions are available. | |
| 2 | View Details | [110..214] | 23.0 | Crystal Structure of HoxA9 and Pbx1 homeodomains bound to DNA | |
| 3 | View Details | [215..283] | 4.066996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
|||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)