













| General Information: |
|
| Name(s) found: |
FTSH1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 716 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
thylakoid lumen [IDA] membrane [IDA] chloroplast thylakoid membrane [IDA] |
| Biological Process: |
photosystem II repair
[TAS]
PSII associated light-harvesting complex II catabolic process [TAS] |
| Molecular Function: |
ATPase activity
[ISS]
protein binding [IPI] ATP-dependent peptidase activity [ISS] metallopeptidase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASNSLLRSS SNFFLGSHII ISSPTPKTTR KPSFPFSFVS RAKYQITRSS QDENSPNGKP 60
61 NSPFSSQVAL AAILLSSISS SPLALAVVDE PASPSVVIES QAVKPSTPSP LFIQNEILKA 120
121 PSPKSSDLPE GSQWRYSEFL NAVKKGKVER VRFSKDGSVV QLTAVDNRRA SVIVPNDPDL 180
181 IDILAMNGVD ISVSEGESSG NDLFTVIGNL IFPLLAFGGL FLLFRRAQGG PGGGPGGLGG 240
241 PMDFGRSKSK FQEVPETGVS FADVAGADQA KLELQEVVDF LKNPDKYTAL GAKIPKGCLL 300
301 VGPPGTGKTL LARAVAGEAG VPFFSCAASE FVELFVGVGA SRVRDLFEKA KSKAPCIVFI 360
361 DEIDAVGRQR GAGMGGGNDE REQTINQLLT EMDGFSGNSG VIVLAATNRP DVLDSALLRP 420
421 GRFDRQVTVD RPDVAGRVKI LQVHSRGKAL GKDVDFDKVA RRTPGFTGAD LQNLMNEAAI 480
481 LAARRELKEI SKDEISDALE RIIAGPEKKN AVVSEEKKRL VAYHEAGHAL VGALMPEYDP 540
541 VAKISIIPRG QAGGLTFFAP SEERLESGLY SRSYLENQMA VALGGRVAEE VIFGDENVTT 600
601 GASNDFMQVS RVARQMIERF GFSKKIGQVA VGGPGGNPFM GQQMSSQKDY SMATADIVDA 660
661 EVRELVEKAY KRATEIITTH IDILHKLAQL LIEKETVDGE EFMSLFIDGQ AELYIS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
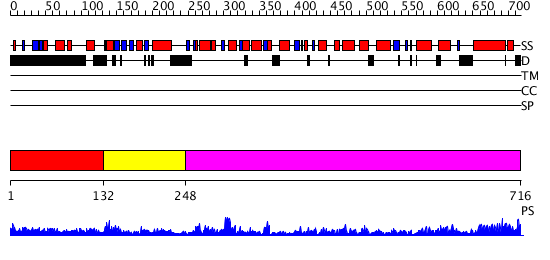
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..131] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [132..247] | 23.180456 | No description for PF06480.6 was found. No confident structure predictions are available. | |
| 3 | View Details | [248..716] | 125.0 | EDTA treated |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.34 |
Source: Reynolds et al. (2008)