













| General Information: |
|
| Name(s) found: |
DRL40_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 888 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
defense response
[ISS]
|
| Molecular Function: |
ATP binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGGCVSVSIS CDQLTKNVCS CLNRNGDYIH GLEENLTALQ RALEQIEQRR EDLLRKILSE 60
61 ERRGLQRLSV VQGWVSKVEA IVPRVNELVR MRSVQVQRLC LCGFCSKNLV SSYRYGKRVM 120
121 KMIEEVEVLR YQGDFAVVAE RVDAARVEER PTRPMVAMDP MLESAWNRLM EDEIGILGLH 180
181 GMGGVGKTTL LSHINNRFSR VGGEFDIVIW IVVSKELQIQ RIQDEIWEKL RSDNEKWKQK 240
241 TEDIKASNIY NVLKHKRFVL LLDDIWSKVD LTEVGVPFPS RENGCKIVFT TRLKEICGRM 300
301 GVDSDMEVRC LAPDDAWDLF TKKVGEITLG SHPEIPTVAR TVAKKCRGLP LALNVIGETM 360
361 AYKRTVQEWR SAIDVLTSSA AEFSGMEDEI LPILKYSYDN LKSEQLKLCF QYCALFPEDH 420
421 NIEKNDLVDY WIGEGFIDRN KGKAENQGYE IIGILVRSCL LMEENQETVK MHDVVREMAL 480
481 WIASDFGKQK ENFIVQAGLQ SRNIPEIEKW KVARRVSLMF NNIESIRDAP ESPQLITLLL 540
541 RKNFLGHISS SFFRLMPMLV VLDLSMNRDL RHLPNEISEC VSLQYLSLSR TRIRIWPAGL 600
601 VELRKLLYLN LEYTRMVESI CGISGLTSLK VLRLFVSGFP EDPCVLNELQ LLENLQTLTI 660
661 TLGLASILEQ FLSNQRLASC TRALRIENLN PQSSVISFVA TMDSLQELHF ADSDIWEIKV 720
721 KRNETVLPLH IPTTTTFFPN LSQVSLEFCT RLRDLTWLIF APNLTVLRVI SASDLKEVIN 780
781 KEKAEQQNLI PFQELKELRL ENVQMLKHIH RGPLPFPCLQ KILVNGCSEL RKLPLNFTSV 840
841 PRGDLVIEAH KKWIEILEWE DEATKARFLP TLKAFPENID ADGYEISF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
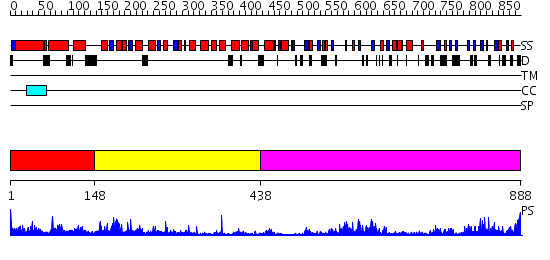
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..147] | 1.03 | Fibrinogen C-terminal domains; Fibrinogen gamma chain | |
| 2 | View Details | [148..437] | 7.154902 | Structure of the apoptotic protease-activating factor 1 bound to ADP | |
| 3 | View Details | [438..888] | 13.69897 | Leucine rich effector protein YopM |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)