













| General Information: |
|
| Name(s) found: |
DNAJ2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 419 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA]
|
| Biological Process: |
protein folding
[ISS]
|
| Molecular Function: |
protein binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFGRGPSRKS DNTKFYEILG VPKTAAPEDL KKAYKKAAIK NHPDKGGDPE KFKELAQAYE 60
61 VLSDPEKREI YDQYGEDALK EGMGGGGGGH DPFDIFSSFF GSGGHPFGSH SRGRRQRRGE 120
121 DVVHPLKVSL EDVYLGTTKK LSLSRKALCS KCNGKGSKSG ASMKCGGCQG SGMKISIRQF 180
181 GPGMMQQVQH ACNDCKGTGE TINDRDRCPQ CKGEKVVSEK KVLEVNVEKG MQHNQKITFS 240
241 GQADEAPDTV TGDIVFVIQQ KEHPKFKRKG EDLFVEHTIS LTEALCGFQF VLTHLDKRQL 300
301 LIKSKPGEVV KPDSYKAISD EGMPIYQRPF MKGKLYIHFT VEFPESLSPD QTKAIEAVLP 360
361 KPTKAAISDM EIDDCEETTL HDVNIEDEMK RKAQAQREAY DDDEEDHPGG AQRVQCAQQ |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
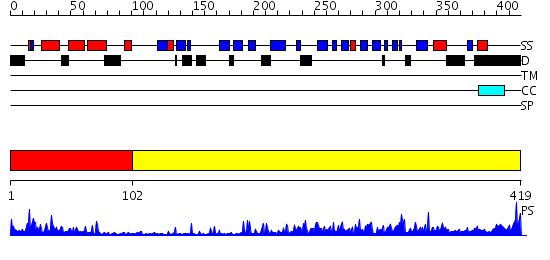
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..101] | 17.69897 | J-DOMAIN (RESIDUES 1-77) OF THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 1-104) OF THE MOLECULAR CHAPERONE DNAJ, NMR, 20 STRUCTURES | |
| 2 | View Details | [102..419] | 47.69897 | The crystal structure of Hsp40 Ydj1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)