













| General Information: |
|
| Name(s) found: |
CPSF2_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 739 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
mRNA cleavage and polyadenylation specificity factor complex [ISS][NAS] |
| Biological Process: |
posttranscriptional gene silencing by RNA
[IMP]
embryonic development ending in seed dormancy [NAS] mRNA polyadenylation [ISS] mRNA cleavage [ISS] |
| Molecular Function: |
DNA binding
[IPI]
protein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGTSVQVTPL CGVYNENPLS YLVSIDGFNF LIDCGWNDLF DTSLLEPLSR VASTIDAVLL 60
61 SHPDTLHIGA LPYAMKQLGL SAPVYATEPV HRLGLLTMYD QFLSRKQVSD FDLFTLDDID 120
121 SAFQNVIRLT YSQNYHLSGK GEGIVIAPHV AGHMLGGSIW RITKDGEDVI YAVDYNHRKE 180
181 RHLNGTVLQS FVRPAVLITD AYHALYTNQT ARQQRDKEFL DTISKHLEVG GNVLLPVDTA 240
241 GRVLELLLIL EQHWSQRGFS FPIYFLTYVS SSTIDYVKSF LEWMSDSISK SFETSRDNAF 300
301 LLRHVTLLIN KTDLDNAPPG PKVVLASMAS LEAGFAREIF VEWANDPRNL VLFTETGQFG 360
361 TLARMLQSAP PPKFVKVTMS KRVPLAGEEL IAYEEEQNRL KREEALRASL VKEEETKASH 420
421 GSDDNSSEPM IIDTKTTHDV IGSHGPAYKD ILIDGFVPPS SSVAPMFPYY DNTSEWDDFG 480
481 EIINPDDYVI KDEDMDRGAM HNGGDVDGRL DEATASLMLD TRPSKVMSNE LIVTVSCSLV 540
541 KMDYEGRSDG RSIKSMIAHV SPLKLVLVHA IAEATEHLKQ HCLNNICPHV YAPQIEETVD 600
601 VTSDLCAYKV QLSEKLMSNV IFKKLGDSEV AWVDSEVGKT ERDMRSLLPM PGAASPHKPV 660
661 LVGDLKIADF KQFLSSKGVQ VEFAGGGALR CGEYVTLRKV GPTGQKGGAS GPQQILIEGP 720
721 LCEDYYKIRD YLYSQFYLL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
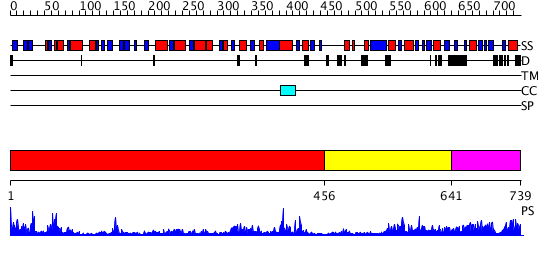
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..455] | 79.0 | No description for 2i7tA was found. | |
| 2 | View Details | [456..640] | 3.32 | No description for 2i7xA was found. | |
| 3 | View Details | [641..739] | N/A | Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)