













| General Information: |
|
| Name(s) found: |
CNGC3_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 706 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA]
|
| Biological Process: |
ion transport
[IEA]
|
| Molecular Function: |
ion channel activity
[ISS]
calmodulin binding [TAS] cyclic nucleotide binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMNPQRNKFV RFNGNDDEFS TKTTRPSVSS VMKTVRRSFE KGSEKIRTFK RPLSVHSNKN 60
61 KENNKKKKIL RVMNPNDSYL QSWNKIFLLL SVVALAFDPL FFYIPYVKPE RFCLNLDKKL 120
121 QTIACVFRTF IDAFYVVHML FQFHTGFITP SSSGFGRGEL NEKHKDIALR YLGSYFLIDL 180
181 LSILPIPQVV VLAIVPRMRR PASLVAKELL KWVIFCQYVP RIARIYPLFK EVTRTSGLVT 240
241 ETAWAGAALN LFLYMLASHV FGSFWYLISI ERKDRCWREA CAKIQNCTHA YLYCSPTGED 300
301 NRLFLNGSCP LIDPEEITNS TVFNFGIFAD ALQSGVVESR DFPKKFFYCF WWGLRNLSAL 360
361 GQNLKTSAFE GEIIFAIVIC ISGLVLFALL IGNMQKYLQS TTVRVEEMRV KRRDAEQWMS 420
421 HRMLPDDLRK RIRKYEQYKW QETKGVEEEA LLSSLPKDLR KDIKRHLCLK LLKKVPWFQA 480
481 MDDRLLDALC ARLKTVLYTE KSYIVREGEP VEDMLFIMRG NLISTTTYGG RTGFFNSVDL 540
541 VAGDFCGDLL TWALDPLSSQ FPISSRTVQA LTEVEGFLLS ADDLKFVATQ YRRLHSKQLR 600
601 HMFRFYSVQW QTWAACFIQA AWKRHCRRKL SKALREEEGK LHNTLQNDDS GGNKLNLGAA 660
661 IYASRFASHA LRNLRANAAA RNSRFPHMLT LLPQKPADPE FPMDET |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
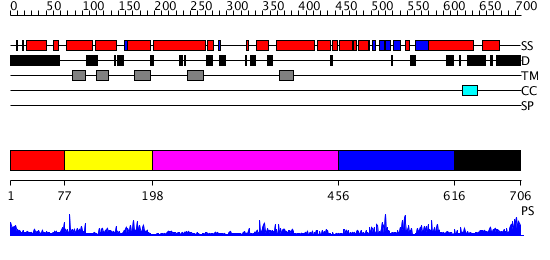
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..76] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [77..197] | 1.53 | Crystal structure of KvAP-33H1 Fv complex | |
| 3 | View Details | [198..455] | 9.09691 | Mammalian Shaker Kv1.2 potassium channel- beta subunit complex | |
| 4 | View Details | [456..615] | 16.0 | Solution structure of the cNMP-binding domain from Arabidopsis thaliana cyclic nucleotide-regulated ion channel | |
| 5 | View Details | [616..706] | 1.876991 | View MSA. No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|