













| General Information: |
|
| Name(s) found: |
CNG18_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 706 amino acids |
Gene Ontology: |
|
| Cellular Component: |
apical plasma membrane
[IDA]
|
| Biological Process: |
pollen tube growth
[IMP]
cellular calcium ion homeostasis [IDA] |
| Molecular Function: |
ion channel activity
[ISS]
calmodulin binding [ISS] cyclic nucleotide binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNKIRSLRCL LPETITSAST AASNRGSDGS QFSVLWRHQI LDPDSNIVTY WNHVFLITSI 60
61 LALFLDPFYF YVPYVGGPAC LSIDISLAAT VTFFRTVADI FHLLHIFMKF RTAFVARSSR 120
121 VFGRGELVMD SREIAMRYLK TDFLIDVAAM LPLPQLVIWL VIPAATNGTA NHANSTLALI 180
181 VLVQYIPRSF IIFPLNQRII KTTGFIAKTA WAGAAYNLLL YILASHVLGA MWYLSSIGRQ 240
241 FSCWSNVCKK DNALRVLDCL PSFLDCKSLE QPERQYWQNV TQVLSHCDAT SSTTNFKFGM 300
301 FAEAFTTQVA TTDFVSKYLY CLWWGLRNLS SYGQNITTSV YLGETLFCIT ICIFGLILFT 360
361 LLIGNMQSSL QSMSVRVEEW RVKRRDTEEW MRHRQLPPEL QERVRRFVQY KWLATRGVDE 420
421 ESILHSLPTD LRREIQRHLC LSLVRRVPFF SQMDDQLLDA ICGCLVSSLS TAGTYIFREG 480
481 DPVNEMLFVI RGQIESSTTN GGRSGFFNST TLRPGDFCGE ELLTWALMPN STLNLPSSTR 540
541 SVRALSEVEA FALSAEDLKF VAHQFKRLQS KKLQHAFRYY SHQWRAWGAC FVQSAWRRYK 600
601 RRKLAKELSL HESSGYYYPD ETGYNEEDEE TREYYYGSDE EGGSMDNTNL GATILASKFA 660
661 ANTRRGTNQK ASSSSTGKKD GSSTSLKMPQ LFKPDEPDFS IDKEDV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
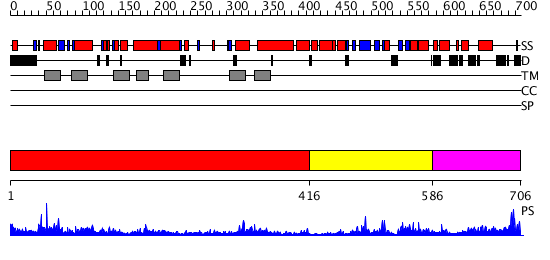
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..415] | 50.522879 | Mammalian Shaker Kv1.2 potassium channel- beta subunit complex | |
| 2 | View Details | [416..585] | 16.0 | Solution structure of the cNMP-binding domain from Arabidopsis thaliana cyclic nucleotide-regulated ion channel | |
| 3 | View Details | [586..706] | 1.774996 | View MSA. No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|