













| General Information: |
|
| Name(s) found: |
CNG16_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 705 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA]
|
| Biological Process: |
ion transport
[IEA]
|
| Molecular Function: |
ion channel activity
[ISS]
calmodulin binding [ISS] cyclic nucleotide binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNLHLYTSA RFRNFPTTFS LRHHHNDPNN QRRRSIFSKL RDKTLDPGGD LITRWNHIFL 60
61 ITCLLALFLD PLYFYLPIVQ AGTACMSIDV RFGIFVTCFR NLADLSFLIH ILLKFKTAFV 120
121 SKSSRVFGRG ELVMDRREIA IRYLKSEFVI DLAATLPLPQ IMIWFVIPNA GEFRYAAHQN 180
181 HTLSLIVLIQ YVPRFLVMLP LNRRIIKATG VAAKTAWSGA AYNLILYLLV SHVLGSVWYV 240
241 LSIQRQHECW RRECIKEMNA THSPSCSLLF LDCGSLHDPG RQAWMRITRV LSNCDARNDD 300
301 DQHFQFGMFG DAFTNDVTSS PFFDKYFYCL WWGLRNLSSY GQSLAASTLS SETIFSCFIC 360
361 VAGLVFFSHL IGNVQNYLQS TTARLDEWRV RRRDTEEWMR HRQLPDELQE RVRRFVQYKW 420
421 LTTRGVDEEA ILRALPLDLR RQIQRHLCLA LVRRVPFFAQ MDDQLLDAIC ERLVPSLNTK 480
481 DTYVIREGDP VNEMLFIIRG QMESSTTDGG RSGFFNSITL RPGDFCGEEL LTWALVPNIN 540
541 HNLPLSTRTV RTLSEVEAFA LRAEDLKFVA NQFRRLHSKK LQHAFRYYSH QWRAWGTCFI 600
601 QAAWRRYMKR KLAMELARQE EEDDYFYDDD GDYQFEEDMP ESNNNNGDEN SSNNQNLSAT 660
661 ILASKFAANT KRGVLGNQRG STRIDPDHPT LKMPKMFKPE DPGFF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
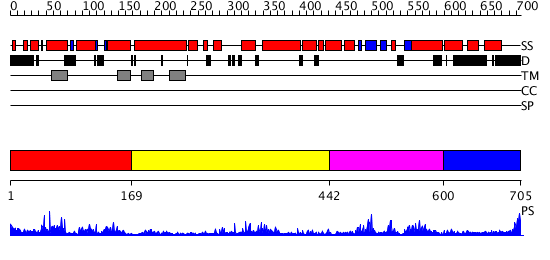
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..168] | 0.95 | Mammalian Shaker Kv1.2 potassium channel- beta subunit complex | |
| 2 | View Details | [169..441] | 16.69897 | Mammalian Shaker Kv1.2 potassium channel- beta subunit complex | |
| 3 | View Details | [442..599] | 16.522879 | Solution structure of the cNMP-binding domain from Arabidopsis thaliana cyclic nucleotide-regulated ion channel | |
| 4 | View Details | [600..705] | N/A | No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|