













| General Information: |
|
| Name(s) found: |
CATA2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 492 amino acids |
Gene Ontology: |
|
| Cellular Component: |
peroxisome
[IDA][NAS]
chloroplast [IDA] cytosolic ribosome [IDA] mitochondrion [IDA] stromule [IDA] |
| Biological Process: |
response to cold
[IEP]
response to oxidative stress [IMP] cell death [IMP] cellular response to sulfate starvation [IEP] cell redox homeostasis [IMP] cellular response to phosphate starvation [IEP] photoperiodism [IMP] response to light stimulus [IEP] hydrogen peroxide catabolic process [NAS] cellular response to nitrogen starvation [IEP] |
| Molecular Function: |
protein binding
[IPI]
catalase activity [IMP][ISS] cobalt ion binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDPYKYRPAS SYNSPFFTTN SGAPVWNNNS SMTVGPRGPI LLEDYHLVEK LANFDRERIP 60
61 ERVVHARGAS AKGFFEVTHD ISNLTCADFL RAPGVQTPVI VRFSTVIHER GSPETLRDPR 120
121 GFAVKFYTRE GNFDLVGNNF PVFFIRDGMK FPDMVHALKP NPKSHIQENW RILDFFSHHP 180
181 ESLNMFTFLF DDIGIPQDYR HMDGSGVNTY MLINKAGKAH YVKFHWKPTC GVKSLLEEDA 240
241 IRVGGTNHSH ATQDLYDSIA AGNYPEWKLF IQIIDPADED KFDFDPLDVT KTWPEDILPL 300
301 QPVGRMVLNK NIDNFFAENE QLAFCPAIIV PGIHYSDDKL LQTRVFSYAD TQRHRLGPNY 360
361 LQLPVNAPKC AHHNNHHEGF MNFMHRDEEV NYFPSRYDQV RHAEKYPTPP AVCSGKRERC 420
421 IIEKENNFKE PGERYRTFTP ERQERFIQRW IDALSDPRIT HEIRSIWISY WSQADKSLGQ 480
481 KLASRLNVRP SI |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
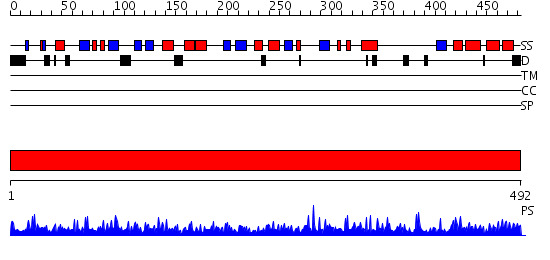
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..492] | 1000.0 | The crystal structure of bovine liver catalase without NADPH |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)