













| General Information: |
|
| Name(s) found: |
CAPP1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 967 amino acids |
Gene Ontology: |
|
| Cellular Component: |
apoplast
[IDA]
cytosol [IDA] |
| Biological Process: |
tricarboxylic acid cycle
[ISS]
|
| Molecular Function: |
protein binding
[IPI]
phosphoenolpyruvate carboxylase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MANRKLEKMA SIDVHLRQLV PGKVSEDDKL VEYDALLLDR FLDILQDLHG EDLRETVQEL 60
61 YEHSAEYEGK HEPKKLEELG SVLTSLDPGD SIVIAKAFSH MLNLANLAEE VQIAYRRRIK 120
121 KLKKGDFVDE SSATTESDLE ETFKKLVGDL NKSPEEIFDA LKNQTVDLVL TAHPTQSVRR 180
181 SLLQKHGRIR DCLAQLYAKD ITPDDKQELD EALQREIQAA FRTDEIKRTP PTPQDEMRAG 240
241 MSYFHETIWK GVPKFLRRVD TALKNIGIEE RVPYNAPLIQ FSSWMGGDRD GNPRVTPEVT 300
301 RDVCLLARMM AATMYFNQIE DLMFEMSMWR CNDELRARAD EVHANSRKDA AKHYIEFWKS 360
361 IPTTEPYRVI LGDVRDKLYH TRERAHQLLS NGHSDVPVEA TFINLEQFLE PLELCYRSLC 420
421 SCGDRPIADG SLLDFLRQVS TFGLSLVRLD IRQESDRHTD VLDAITTHLD IGSYREWSEE 480
481 RRQEWLLSEL SGKRPLFGSD LPKTEEIADV LDTFHVIAEL PADSFGAYII SMATAPSDVL 540
541 AVELLQRECR VKQPLRVVPL FEKLADLEAA PAAVARLFSV DWYKNRINGK QEVMIGYSDS 600
601 GKDAGRLSAA WQLYKAQEEL VKVAKEYGVK LTMFHGRGGT VGRGGGPTHL AILSQPPDTI 660
661 NGSLRVTVQG EVIEQSFGEE HLCFRTLQRF TAATLEHGMR PPISPKPEWR ALLDEMAVVA 720
721 TEEYRSVVFQ EPRFVEYFRL ATPELEYGRM NIGSRPSKRK PSGGIESLRA IPWIFAWTQT 780
781 RFHLPVWLGF GSAIRHVIEK DVRNLHMLQD MYQHWPFFRV TIDLIEMVFA KGDPGIAALY 840
841 DKLLVSEELW PFGEKLRANF EETKKLILQT AGHKDLLEGD PYLKQRLRLR DSYITTLNVC 900
901 QAYTLKRIRD PSYHVTLRPH ISKEIAESSK PAKELIELNP TSEYAPGLED TLILTMKGIA 960
961 AGLQNTG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
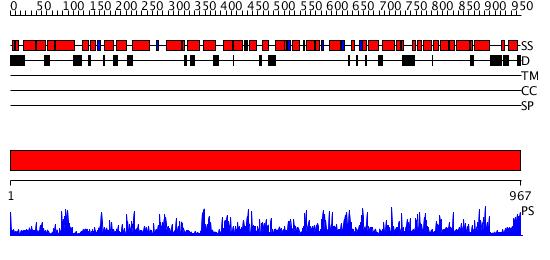
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..967] | 1000.0 | Phosphoenolpyruvate carboxylase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)