













| General Information: |
|
| Name(s) found: |
CAP2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 653 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
plasma membrane [IDA] |
| Biological Process: |
clathrin coat assembly
[IEA]
|
| Molecular Function: |
clathrin binding
[IEA]
phospholipid binding [IEA] binding [ISS] phosphatidylinositol binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAPSIRKAIG AVKDQTSIGI AKVASNMAPD LEVAIVKATS HDDDPASEKY IREILNLTSL 60
61 SRGYILACVT SVSRRLSKTR DWVVALKALM LVHRLLNEGD PIFQEEILYS TRRGTRMLNM 120
121 SDFRDEAHSS SWDHSAFVRT YAGYLDQRLE LALFERKSGV SVNSGGNSSH HSNNDDRYGR 180
181 GRDDFRSPPP RSYDYENGGG GGSDFRGDNN GYGGVPKRSR SYGDMTEMGG GGGGGGRDEK 240
241 KVVTPLREMT PERIFGKMGH LQRLLDRFLS LRPTGLAKNS RMILIALYPV VRESFKLYAD 300
301 ICEVLAVLLD KFFDMEYSDC VKAFDAYASA AKQIDELIAF YNWCKETGVA RSSEYPEVQR 360
361 ITSKLLETLE EFVRDRAKRG KSPERKEIEA PPPVVEEEEP EPDMNEIKAL PPPENYTPPP 420
421 PPEPEPQPEK PQFTEDLVNL REDEVTADDQ GNKFALALFA GPPGNNGKWE AFSSNGVTSA 480
481 WQNPAAEPGK ADWELALVET TSNLEKQTAA LGGGFDNLLL NGMYDQGMVR QHVSTSQLTG 540
541 GSASSVALPL PGKTNNQVLA LPAPDGTVEK VNQDPFAASL TIPPPSYVQM AEMEKKQYLL 600
601 SQEQQLWQQY QRDGMRGQAS LAKMNTGPVP AYGMPPVNGM GPPPTGYYYN NPY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
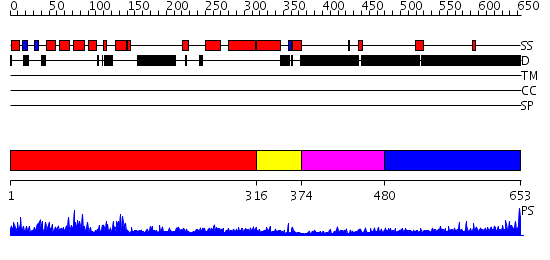
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..315] | 89.0 | Clathrin assembly lymphoid myeloid leukaemia protein, Calm | |
| 2 | View Details | [316..373] | 2.45 | Clathrin assembly lymphoid myeloid leukaemia protein, Calm | |
| 3 | View Details | [374..479] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [480..653] | 5.69897 | Sec24 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)