













| General Information: |
|
| Name(s) found: |
BSU1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 793 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
regulation of protein localization
[IDA]
brassinosteroid mediated signaling pathway [IGI] |
| Molecular Function: |
protein serine/threonine phosphatase activity
[IDA][ISS]
protein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAPDQSYQYP SPSYESIQTF YDTDEDWPGP RCGHTLTAVF VNNSHQLILF GGSTTAVANH 60
61 NSSLPEISLD GVTNSVHSFD VLTRKWTRLN PIGDVPSPRA CHAAALYGTL ILIQGGIGPS 120
121 GPSDGDVYML DMTNNKWIKF LVGGETPSPR YGHVMDIAAQ RWLVIFSGNN GNEILDDTWA 180
181 LDTRGPFSWD RLNPSGNQPS GRMYASGSSR EDGIFLLCGG IDHSGVTLGD TYGLKMDSDN 240
241 VWTPVPAVAP SPRYQHTAVF GGSKLHVIGG ILNRARLIDG EAVVAVLDTE TGEWVDTNQP 300
301 ETSASGANRQ NQYQLMRRCH HAAASFGSHL YVHGGIREDV LLDDLLVAET SQSSSPEPEE 360
361 DNPDNYMLLD DYLMDEPKPL SSEPEASSFI MRSTSEIAMD RLAEAHNLPT IENAFYDSAI 420
421 EGYVPLQHGA ETVGNRGGLV RTASLDQSTQ DLHKKVISTL LRPKTWTPPA NRDFFLSYLE 480
481 VKHLCDEVEK IFMNEPTLLQ LKVPIKVFGD IHGQYGDLMR LFHEYGHPSV EGDITHIDYL 540
541 FLGDYVDRGQ HSLEIIMLLF ALKIEYPKNI HLIRGNHESL AMNRIYGFLT ECEERMGESY 600
601 GFEAWLKINQ VFDYLPLAAL LEKKVLCVHG GIGRAVTIEE IENIERPAFP DTGSMVLKDI 660
661 LWSDPTMNDT VLGIVDNARG EGVVSFGPDI VKAFLERNGL EMILRAHECV IDGFERFADG 720
721 RLITVFSATN YCGTAQNAGA ILVIGRDMVI YPKLIHPHPP PISSSEEDYT DKAWMQELNI 780
781 EMPPTPARGE SSE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
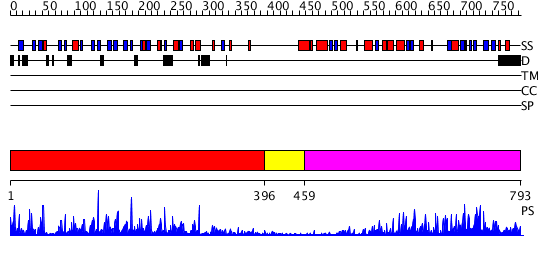
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..395] | 56.39794 | No description for 2vpjA was found. | |
| 2 | View Details | [396..458] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [459..793] | 94.154902 | Protein phosphatase-1 (PP-1) |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)