













| General Information: |
|
| Name(s) found: |
ARP_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 536 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast nucleoid
[IDA]
|
| Biological Process: |
positive regulation of transcription
[TAS]
|
| Molecular Function: |
DNA-(apurinic or apyrimidinic site) lyase activity
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNNVLQFGLQ SSAIYVAKFL VVPLRSLRVG SSFVGVGVGT RSFNKRLMSN ATAFSINNSK 60
61 RKELKIPGAA IDQNCHQMGS DTDRDEMGTL QDDRKEIEAM TVQELRSTLR KLGVPVKGRK 120
121 QELISTLRLH MDSNLPDQKE TSSSTRSDSV TIKRKISNRE EPTEDECTNS EAYDIEHGEK 180
181 RVKQSTEKNL KAKVSAKAIA KEQKSLMRTG KQQIQSKEET SSTISSELLK TEEIISSPSQ 240
241 SEPWTVLAHK KPQKDWKAYN PKTMRPPPLP EGTKCVKVMT WNVNGLRGLL KFESFSALQL 300
301 AQRENFDILC LQETKLQVKD VEEIKKTLID GYDHSFWSCS VSKLGYSGTA IISRIKPLSV 360
361 RYGTGLSGHD TEGRIVTAEF DSFYLINTYV PNSGDGLKRL SYRIEEWDRT LSNHIKELEK 420
421 SKPVVLTGDL NCAHEEIDIF NPAGNKRSAG FTIEERQSFG ANLLDKGFVD TFRKQHPGVV 480
481 GYTYWGYRHG GRKTNKGWRL DYFLVSQSIA ANVHDSYILP DINGSDHCPI GLILKL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
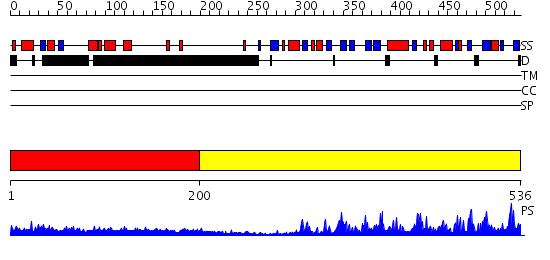
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..199] | 11.176996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [200..536] | 81.39794 | DNA repair endonuclease Hap1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)