













| General Information: |
|
| Name(s) found: |
ADR1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 787 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endomembrane system
[IEA]
|
| Biological Process: |
response to other organism
[IEP]
defense response [ISS] response to water deprivation [IMP] |
| Molecular Function: |
kinase activity
[TAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASFIDLFAG DITTQLLKLL ALVANTVYSC KGIAERLITM IRDVQPTIRE IQYSGAELSN 60
61 HHQTQLGVFY EILEKARKLC EKVLRCNRWN LKHVYHANKM KDLEKQISRF LNSQILLFVL 120
121 AEVCHLRVNG DRIERNMDRL LTERNDSLSF PETMMEIETV SDPEIQTVLE LGKKKVKEMM 180
181 FKFTDTHLFG ISGMSGSGKT TLAIELSKDD DVRGLFKNKV LFLTVSRSPN FENLESCIRE 240
241 FLYDGVHQRK LVILDDVWTR ESLDRLMSKI RGSTTLVVSR SKLADPRTTY NVELLKKDEA 300
301 MSLLCLCAFE QKSPPSPFNK YLVKQVVDEC KGLPLSLKVL GASLKNKPER YWEGVVKRLL 360
361 RGEAADETHE SRVFAHMEES LENLDPKIRD CFLDMGAFPE DKKIPLDLLT SVWVERHDID 420
421 EETAFSFVLR LADKNLLTIV NNPRFGDVHI GYYDVFVTQH DVLRDLALHM SNRVDVNRRE 480
481 RLLMPKTEPV LPREWEKNKD EPFDAKIVSL HTGEMDEMNW FDMDLPKAEV LILNFSSDNY 540
541 VLPPFIGKMS RLRVLVIINN GMSPARLHGF SIFANLAKLR SLWLKRVHVP ELTSCTIPLK 600
601 NLHKIHLIFC KVKNSFVQTS FDISKIFPSL SDLTIDHCDD LLELKSIFGI TSLNSLSITN 660
661 CPRILELPKN LSNVQSLERL RLYACPELIS LPVEVCELPC LKYVDISQCV SLVSLPEKFG 720
721 KLGSLEKIDM RECSLLGLPS SVAALVSLRH VICDEETSSM WEMVKKVVPE LCIEVAKKCF 780
781 TVDWLDD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
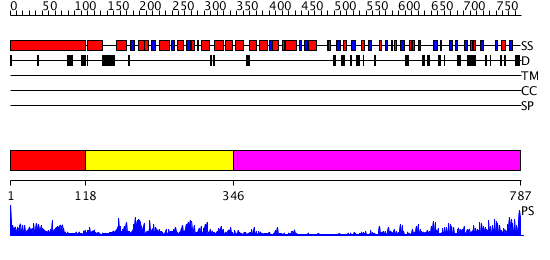
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..117] | 2.735996 | View MSA. Confident ab initio structure predictions are available. | |
| 2 | View Details | [118..345] | 10.154902 | Structure of the apoptotic protease-activating factor 1 bound to ADP | |
| 3 | View Details | [346..787] | 10.09691 | No description for 2z7xB was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)