













| General Information: |
|
| Name(s) found: |
DNAJ_BACSU
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Bacillus subtilis |
| Length: | 372 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKRDYYEVL GVSKSASKDE IKKAYRKLSK KYHPDINKEA GSDEKFKEVK EAYETLSDDQ 60
61 KRAHYDQFGH TDPNQGFGGG GFGGGDFGGF GFDDIFSSIF GGGTRRRDPK LRARGADLQY 120
121 TMTLSFEDAA FGKETTIEIP REETCETCKG SGAKPGTNPE TCSHCGGSGQ LNVEQNTPFG 180
181 KVVNRRVCHH CEGTGKIIKN KCADCGGKGK IKKRKKINVT IPAGVDDGQQ LRLSGQGEPG 240
241 INGGLPDLFV VFHVRAHEFF ERDGDDIYCE MPLTFAQAAL GDEVEVPTLH GKVKIPAGTQ 300
301 TGTKFRLRGK GVQNVRGYGQ GDQHIVVRVV TPTNLTDKQK DIIREFAEVS GNLPDEQEMS 360
361 FFDKVKRAFK GD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
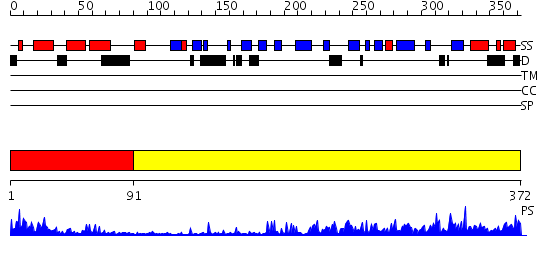
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..90] | 19.69897 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 2 | View Details | [91..372] | 45.221849 | The crystal structure of Hsp40 Ydj1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)