













| General Information: |
|
| Name(s) found: |
gbh-2 /
CE35896
[WormBase]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 409 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell
[IDA]
mitochondrion [TAS] |
| Biological Process: |
electron transport
[IEA carnitine biosynthetic process [IEA |
| Molecular Function: |
L-ascorbic acid binding
[IEA trimethyllysine dioxygenase activity [IEA oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen [ISS] oxidoreductase activity [IEA iron ion binding [IEA oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIVQRRLVSG FTWVTGWNLK NIRSKNDCLE IRYENEGNPS KLIMPFVWLR DHCTSQKLYH 60
61 LPTNQRKSNC CDITSLSKIK HSNQVIIDEA TNSLQIVWID GHQSKFKIGN IIREGKVEKN 120
121 VSNDNKIYEL WNSKSLKDVP RISKSTLSLQ SFSKNLVKYG VIIVDGVEGT SEATEKLCQS 180
181 LVPVHDTFFG QFWVFSNSAT NDEPAYEDTA YGSDEIGPHT DGTYFDQTPG IQVFHCLTPA 240
241 KTGGDTVLVD SFYCAEKLRN ESPEDFEILC NTKISHHYLE GSPPGSSIHS VSLEKPVIER 300
301 NSFGNITQIR FNPYDRAPFS CLNSSEASAA ETIKFYEAYE KFSKICHNPD NSIEISLRPG 360
361 SVIFIDNFRI LHSRTSFQGY RQMCGCYLSR DNFMAKAIPF LSGKSQTSL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
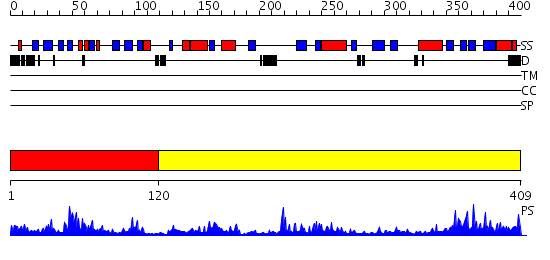
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..119] | 6.206996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [120..409] | 54.522879 | Taurine/alpha-ketoglutarate dioxygenase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)