













| General Information: |
|
| Name(s) found: |
coh-3 /
CE35852
[WormBase]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 607 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear chromosome
[IEA |
| Biological Process: |
embryonic development ending in birth or egg hatching
[IMP]
meiotic chromosome segregation [IMP] meiosis [IMP] |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVISIDVPLK ASEEATVKFL LGIVFGSHKK LGLQSNRREC FRHDVLQASK LIQWILSNER 60
61 KNRRSLSIAC NLVYGNTIVL STQVARLLQD AIRAREIVAF TSYLAEEDER KRKTREQMEG 120
121 DNSKTPKKKR RTMVEEFPLT PTPISVALKQ NITMPDVLSL HKNDQINVED DLAAPTMEQL 180
181 EMMYGMLESE NRNTAIDLTF IDRYSSNDCR QNVHNLNLET DFQRPSDDFI RSQLMTPTKS 240
241 IPQEEEFQPA THLKEMLLFR QLPITLDETE RRRSMEEEME EARNSGINTE FIPPQQMDVP 300
301 RLVSENMDLD DEVSIPIRIP KVPLSATKKQ TSENEGAEEG INYKIPLREM HHIMEDYSSL 360
361 HFKDPEPLYR KTSKVMNLKE LLNPIPYFLR KGCNTDMWDL FKVKTRKFDS TIGHDVSDTE 420
421 EEDEDEETER RNRLKAELEP LETIDLSQMK FMTTPLKDKS PILTPVHRPE EIEFREELEL 480
481 PRFEQFEPMN VNPISPKMEE PALFEDNWNS KPTWSNTSSE NIADALTIRG NIIRQLDDKS 540
541 PFEVTLDSMI PVAATSRREA ARTFYTVLEL LKERKIKATQ RAPYENIDLL LSTDDSEDID 600
601 DLAMADF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
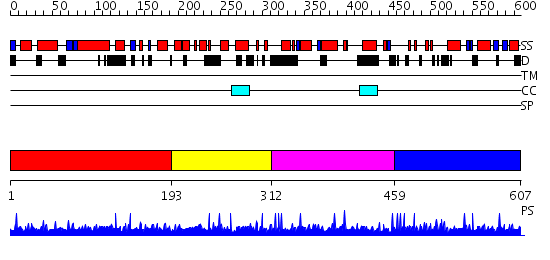
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..192] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [193..311] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [312..458] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [459..607] | 1.53 | Sc Smc1hd:Scc1-C complex, ATPgS |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)