













| General Information: |
|
| Name(s) found: |
CE33911
[WormBase]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 708 amino acids |
Gene Ontology: |
|
| Cellular Component: |
heterotrimeric G-protein complex
[IEA |
| Biological Process: |
G-protein coupled receptor protein signaling pathway
[IEA embryonic development ending in birth or egg hatching [IMP |
| Molecular Function: |
signal transducer activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSKQQIGVL KRDGIGKQYF DMENILTRSL YRVLNSSTQV PPHIVTKSQN VDKMSFPDRK 60
61 SFQTIAVARP NNHQIGNASS VDMDMQMRLL AQEICGRVER KYERKLEDVR ERDRRDTERR 120
121 LISIRSEYES KYQLQLGKLE KEKNRLLDQI AERERTLEHR MEMRSAEREH DLVAKQRTVE 180
181 NRAIELTLNK ENFEKCRDEW QRKMELEMEE LRLERERIEV ASLKSTDKRR SDMAIEAEIS 240
241 MWKKRTAELE HDANETRKRV GEMMEDNFRL KNQISSVGQI QKELDVTMTS LNETREELAV 300
301 SRVEVRRTGD YDQLKEENDQ LRIEIELLRQ KTSQKVKIAV EETIAEYSAK EAKWKRIAGL 360
361 SQQRIAVLSE KLKDSEIERD VLRQEMKTMQ KMVGRSSAGR QGHNQKNTES RKIIRSISPS 420
421 GSTSSLSDGE LEILNIRQRI QNLDDIAKEL DASVEHFSTG GASRKLFDKN ETNVELYDDF 480
481 CRALHSSVMD EEGSPLHMTS SPQKPTQNHK RKIEIPVNEK SMVSTSQVTD QDEWLKRGIR 540
541 VISPPAMKRS PVVRPESPEV ILESEKIEVK IEQPPVKTIE PELTEFEKRM QARSAAKDDT 600
601 DRRNRILQLA TDPETSIVKP KSPEPTEEPA KPTSADNPMF AGVDPVMAEY MKKVLAARNQ 660
661 NQTTLSAQPT IQVAPVVVPP KSTEVEVTLA DELDLELDQP TNADDDWW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
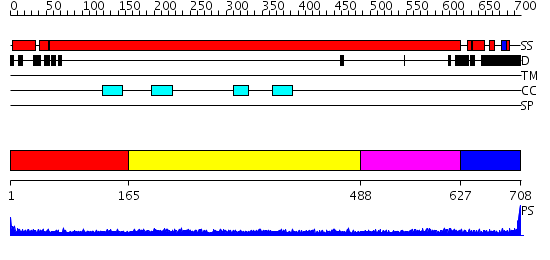
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..164] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [165..487] | 18.0 | Heavy meromyosin subfragment | |
| 3 | View Details | [488..626] | 4.154902 | No description for 2dfsA was found. | |
| 4 | View Details | [627..708] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)