













| General Information: |
|
| Name(s) found: |
CE33608
[WormBase]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 727 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
cell cycle arrest
[IEA |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKFAASGGK RLEEEIGKLQ ADADHIFEAL AVISKDEALQ ESPSLLLVSK QVSKKIEQLK 60
61 VLLKLVEEKK DTFISKCKLV MRTAGPESDA KELDSLMKHV EDAYSDVFEL ISDKEGLIDR 120
121 FNASNINSDS VPNTESLNLA FAAADPEFGA GRLLDAEDVD TNNNPDEKPV ITYVSSLNNA 180
181 PQDPEMSRLA ASGGKGFEEE IGKLQADADH IFEALAVMSK DEALQESPSL LLYSKQIRFN 240
241 KIQKLKELLK PVEEKKDTFI SECKLVMKTA GPEADTKELD SLLKQVEDAY SDVFELISDK 300
301 EGLIDRCNAS NINSDSVSNT ERLNIALAAA DAEFGAGRLL DAEDVDTNNN PDEKSIITYV 360
361 SSLNNAFPLE PEMSKYVASG GKRLEEEIGK LQADADHIFK ALEEISKDEA LQESPSLLLD 420
421 SKQVSKKIEQ LKVLLKPVEE KKDTFISESK LVMKTVGPES DAKELDSLMK HVEDAYSDVV 480
481 ELISDKEGLI DRCNASNRIA HPQTETEKIT NEIELLKETC SCCTPYQIEK ISENYYRFGD 540
541 THIKRMVRIL RSTVMVRVGG GWEPLEESLQ KHDPCRTKGR LNINMFPEAR PIHALDLMQS 600
601 FTKNRHAKQM PIAGTPGPIM KIREKTERSV PMSGGLGGTA GYTVTTESPR KTPQRPFRIP 660
661 RTPSDKTPGR QSRVGCVSNL KNSIVEPSSP SQSESRASPE VADRQKRVSL SLKEKGKRFK 720
721 KNAERAK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
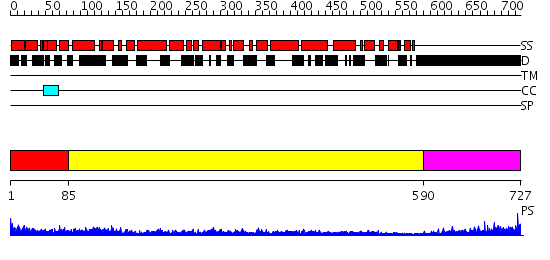
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..84] | 7.0 | Crystal structure of the native monooxygenase domain of MICAL at 1.45 A resolution | |
| 2 | View Details | [85..589] | 53.69897 | Cryo-EM Structure of Chicken Gizzard Smooth Muscle alpha-Actinin | |
| 3 | View Details | [590..727] | 6.128996 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)