













| General Information: |
|
| Name(s) found: |
nuo-5 /
CE33341
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 634 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA formate dehydrogenase complex [IEA |
| Biological Process: |
electron transport
[IEA nematode larval development [IMP positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP ATP synthesis coupled electron transport [IEA formate metabolic process [IEA growth [IMP |
| Molecular Function: |
oxidoreductase activity
[IEA electron carrier activity [IEA molybdenum ion binding [IEA NADH dehydrogenase (ubiquinone) activity [IEA formate dehydrogenase activity [IEA dimethyl sulfoxide reductase activity [IEA oxidoreductase activity, acting on NADH or NADPH [IEA 4 iron, 4 sulfur cluster binding [IEA iron-sulfur cluster binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHRVGGQIVR GVSKAQQKRS LSVAAPAPPK KVEVFIDDKK VLVDPGMTIL QACALVGVDI 60
61 PRFCYHDRLS IAGNCRMCLV EVEKSVKPVA SCAMPVMNGM KVKTNSDFVK KAREGVMEFM 120
121 LNNHPLDCPI CDQGGECDLQ DQAMNFGSDR GRLQSRFDGK RALEDKNIGP LVKTVMTRCI 180
181 QCTRCVRFAN EVAAFPDFGT TGRGQDLQIG TYVEKFFASE LSGNIIDICP VGALTSKQYA 240
241 FTARPWETRK TESVDVMDAT GSNIVLSHRT GELLRVIPKI NDDINEEWIG DQSRFAVDGL 300
301 KVQRLLTPMI RGADGQLKPA TWEEALFTVA AKLRETPAEQ KAAVAGGLND VESLVALKDL 360
361 FNRFNSENVM TEEEFPDVGS GGSDLRSNYV FNDGIASVEN ADAILLVGTN PRFEAPTLNA 420
421 RIRKSFLYSD VQVGVIGAET ELTYEYDYLG ASAKAIDEIL AGKGDFAKIL SSATTPLIIV 480
481 GAQALKGEAG AALLGKLQQL ADKLGSGKEV KVLNVLQRWA GQAGALDVGY KAGTAGIRKT 540
541 PIKFLYLLGA DEGKVTKANL DPSAFVVYQG HHGDAGAEMA DVVLPGAAYT EKEGTYVNTE 600
601 GRSQRCASGK AKKKLDLSIF WQRKSNNFGF FQRK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
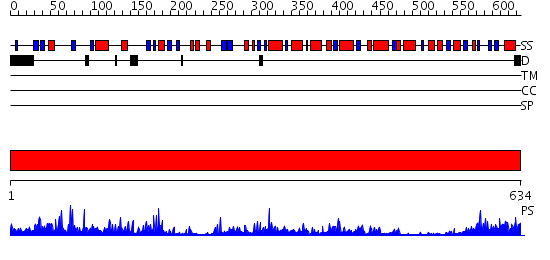
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..634] | 96.0 | Crystal structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)