













| General Information: |
|
| Name(s) found: |
cogc-3 /
CE33242
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 477 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cis-Golgi network
[IEA membrane [IEA |
| Biological Process: |
hermaphrodite genitalia development
[IMP locomotory behavior [IMP morphogenesis of an epithelium [IMP nematode larval development [IMP gonad morphogenesis [IMP embryonic development ending in birth or egg hatching [IMP regulation of cell migration [IMP intracellular protein transport [IEA positive regulation of multicellular organism growth [IMP growth [IMP |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLAECQYAYF AKRNLLLGPI LESTLANLSG THDDSTCRLT RDACTFMLRT CDDEYRLYRQ 60
61 FFVTRHTEER KLSTDGRISP AMSTITSVFT SAQSPQQVHP FETFSEQMCR TLYDMLRPRI 120
121 VHNPHLETLA ELCTMIKIEM IENRCSLQMV ASILGDDASN DQNVSGLNPR AGFVSVMSEL 180
181 VGDIAERIVY RAGMYAQNDI AAYRPAAGDI AYPQMLQMIR KIENEQKEQQ EKEPEEGSAP 240
241 ATAIDQHCLW YPTVRRTVMC LSKIFPCLDI GVFHSLARDM LFSCIDSLET ASKAIMESPA 300
301 PAKGWSKKLD AHLFVVKHLL ILREQTAPYR QNVLSFRSDS LNTKDFSIDF SKFTNVLFDS 360
361 KSKWFELSTD NALLELITTV PIEMREQEGD SRRVLDQQLR ASTFRLAHEA SLLMIRDLAD 420
421 WIEIAEDERM KEGFQLSGHP KLAAGVLKDV SAVAYRNVGT KFTEVNIYSN STVLYII |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
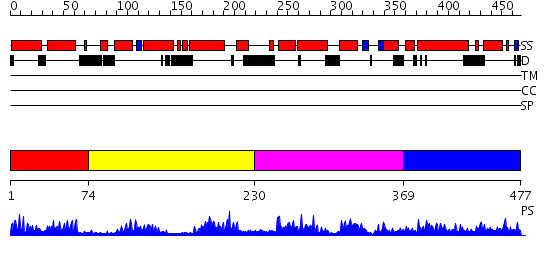
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..73] | 1.024981 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [74..229] | 1.02797 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [230..368] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [369..477] | 2.029982 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)