













| General Information: |
|
| Name(s) found: |
lsd-1 /
CE33174
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 737 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA integral to membrane [IEA |
| Biological Process: |
electron transport
[IEA porphyrin biosynthetic process [IEA |
| Molecular Function: |
oxidoreductase activity
[IEA oxygen-dependent protoporphyrinogen oxidase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASGTRRKAN DDSQSSSEGS SSTTPQFIPQ VPFDFNLGSI NEEVDDSNGL TGIASVPKDI 60
61 LTPLRDHYSL AEVARRHGIS ADRPTEIEAA FFPEVQMSRS FSDVFLMIRN TTLSIWLASA 120
121 TTECTAEDVI KHLTPPYNTE IHLVQNIVLF LSRFGMINIG FFFPKTELVN NMEKKFVVVI 180
181 GAGAAGIAAA TQLLTFGFDV AVVEASGLTG GRVRSLISKH GELIETGCDS LRNLDESVIT 240
241 TLLHQVPLNE NIMSENTIVF SKGKYVPVAR CHVINGLYAN LKAGLAHASH GPEQRGENGL 300
301 YISRQQAYEN YFNMIERSTL LSYYNFAKEK VNLNAERKHL YEVLKTNRLT ALLAEQKLKN 360
361 TPPSDELLLK SLQIDIEKAI RQFDEACERF EICEERIADL EKNPRCKQSM HPNDFIHYNF 420
421 LLGFEERLFG AQLEKVQFSC NVNELKLKSQ VARVQEGLAQ VLINVANERK VKIHHNQRVI 480
481 EIDTGSSDAV ILKLRKPDGS VGILNADYVV STLPIGVLKK TIIGDERAPV FRPPLPKSKF 540
541 AAIRSLGNGL INKIVFVFET RFWPESINQF AIVPDKISER AAMFTWSSLP ESRTLTTHYV 600
601 GENRFHDTPV TELITKALEM LKTVFKDCPS PIDAYVTNWH TDELAFGTGT FMSLRTEPQH 660
661 FDALKEPLKT RDGKPRVFFA GEHTSALEHG TLDGAFNSGL RAAADLANTC IEIPFINRSR 720
721 DLIFPPYPRA RYSNEGD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
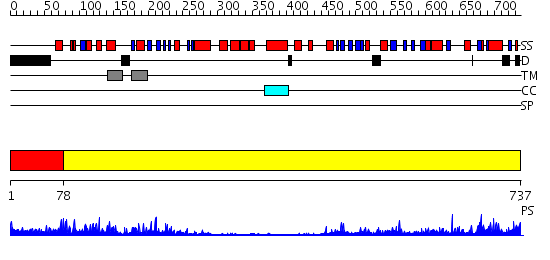
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..77] | 19.09691 | Dihydropyrimidine dehydrogenase, N-terminal domain; Dihydropyrimidine dehydrogenase, domain 4; Dihydropyrimidine dehydrogenase, domain 3; Dihydropyrimidine dehydrogenase, domain 2; Dihydropyrimidine dehydrogenase, C-terminal domain | |
| 2 | View Details | [78..737] | 103.0 | No description for 2v1dA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.41 |
Source: Reynolds et al. (2008)