













| General Information: |
|
| Name(s) found: |
CE33034
[WormBase]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 740 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
locomotory behavior
[IMP nematode larval development [IMP positive regulation of growth rate [IMP purine ribonucleoside monophosphate biosynthetic process [IEA growth [IMP |
| Molecular Function: |
AMP deaminase activity
[IEA adenosine deaminase activity [IEA deaminase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKTVDSELFD SGPFKKNFAF GNADHSTASP FEISTTPIEV NESRAHKTIE ISNRARLESQ 60
61 NDGCLSPSSL KSVLDEIQTE PRCKSPQHID LHTFRDAVDV NYQRMAITGE ELSGVPLEDL 120
121 KTASGHLIEA LHLRSKYMER IGNQFPSTTR NFLSGHYPAN LPKHRVKNTE TTVQTSFNPP 180
181 DPPKDHWGKN DPLPKYEKIY HLRRNRGVTE ICNDDGSIDQ QFKNVNVTKE EFLNDTEKLT 240
241 AMIVDGPLKS FCFRRLSYLE NKFQLHVLLN ELRELHEQKG VSHRDFYNIR KVDTHIHAAS 300
301 SMNQKHLLRF IKKKIKTEAD TVVLNNNGTK VTMKEVFKKM GIDAYDLSVD MLDVHADRNT 360
361 FHRFDKFNTK YNPVGESTLR EIFIKTDNYV GGKYFADLLK EVLSDLEDSK YQHAEPRLSI 420
421 YGRSKNEWDN LAKWALTHDV WSPNARWLVQ IPRLYDVYRA KNMVKNFDDM LDNLFTPLFE 480
481 VTNDPSSHPE LHLFLQQISG IDSVDDESKH EFVNFDRSTP CPPEYTDLEN PPYNYYLFYM 540
541 YRNICALNAF RRARGLNTFA LRPHCGEAGH VSHLLTGYLT SESIAHGILL RKVPVLQYLY 600
601 YLTQIGIAMS PLSNNSLFIS YQRNPLPEYL QKGLNVSLST DDPLQFHYTK EALMEEFSIA 660
661 AQVWKLSSCD MCELARNSVM QSGFEDKVKI HWLGPNYKEE GVLGNDIHRT NVPDIRVSFR 720
721 HEALVDELYN LFRVQNTLKQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
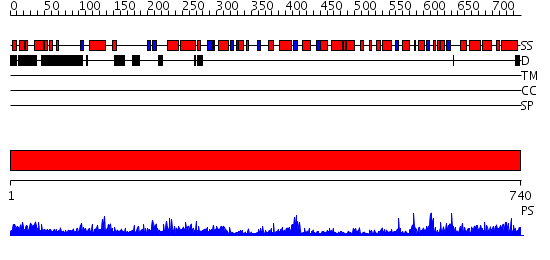
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..740] | 161.0 | X-Ray Structure of Adenosine 5'-Monophosphate Deaminase from Arabidopsis Thaliana in Complex with Coformycin 5'-Phosphate |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)