













| General Information: |
|
| Name(s) found: |
sop-2 /
CE32334
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 735 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
oviposition
[IMP |
| Molecular Function: |
identical protein binding
[IPI]
enzyme binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSNLTSNEM SSTSAIVEPP EAVKGNERDK STRRSTSQVV RPEKHEVENA LEDQCSSSSL 60
61 PKEAQYYAKL DKKLEGKDPR SQFYEAVRLS ADIFAHKFEK AVCSRQTFEP TNSIIKVLNT 120
121 AEEEMLHEKV VPLPVSSKLQ YYLNRGRYDT IFDRDEQLQR TADPMADEPD HVEKRVTSIL 180
181 EQASREMEEG EVEPVFYDGS DEDQELPIDL GAMRNLQRTN KFAARSSRMA ARRGGRPGYR 240
241 GAFRGAARGA PSRRPAPAAE VAPETPVAAP MAPAAPAAPA TPEAAPAAEV MDTSIATEMP 300
301 QESAVDLSNV SAATEMDTSK EGEASRPTSE KKKKIRTTEM DRLMSMDLGP KDGGRVGELG 360
361 HMWPESRRRP AAPLPETPAA QPRKSLPRRA AEKKKPEDSD AAEEQEVEME VDNDASTSTP 420
421 RNARGGRGGG NRRGSRRGQK RTSGGSGKLV EPKKEPVDEP AEKIPKRSEA APEVPATATT 480
481 KEAPPSTSSS PPDAPATPAT PASSDSRDSP RKIRAMIFSL TGSPPESETP PVLQQEQVIS 540
541 TAAPTAGRHP NIIQQVPHIN RIPPQPLRRL TAPQAPPASQ PEEPPVQQTV PVVKVELASA 600
601 PAPIVRDPQS TEPVPPAMPT LVENNHEATL ILPPNKTSDY TRWNAQDLIN WVRLLITNNV 660
661 DSTIAIMVRE EFDGETLACL VLDDDFRKEV PIPYGHYKKM KIYGTEVLNH YRTEKYQADL 720
721 RKFHEELAAW KAQQR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
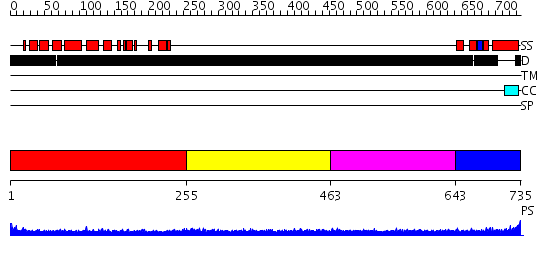
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..254] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [255..462] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [463..642] | 3.186996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [643..735] | N/A | Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)