













| General Information: |
|
| Name(s) found: |
cat-2 /
CE32103
[WormBase]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 393 amino acids |
Gene Ontology: |
|
| Cellular Component: |
axon
[IDA |
| Biological Process: |
response to food
[IMP mechanosensory behavior [IMP dopamine biosynthetic process [IMP serotonin biosynthetic process [IEA dopamine metabolic process [ISS habituation [IMP aromatic amino acid family metabolic process [IEA L-phenylalanine catabolic process [IEA regulation of locomotion [IMP catecholamine biosynthetic process [IEA |
| Molecular Function: |
iron ion binding
[IEA phenylalanine 4-monooxygenase activity [IEA monooxygenase activity [IEA tyrosine 3-monooxygenase activity [IEA tryptophan 5-monooxygenase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFLRYMRRRG SGEDHEEEPR GVTVIATKVA ENSKNPRRYS LVHQASCETQ HHKGIRRQNT 60
61 IQHRKQLTDQ MRCQKILQQL NDEGIEVIFT ANDVTPIEFS IILTSTDPTL SNFVSDILQN 120
121 MSSAKVQICH VETRGNEASH DVLLACKATK NQLIHSAELL TQNHVALTKF SIFAKKLSDE 180
181 KNQSQIWFPR HISELDQCSK CITKYEPTTD PRHPGHGDVA YIARRKFLND QALEFKFGDE 240
241 IGYVDYTEEE HATWKAVYEK LGDLHLSHTC AVYRQNLKIL QEEKVLTADR IPQIRDVNKF 300
301 LQKKTGFELR PCSGLLSARD FLASLAFRVF QTTTYLRHHK SPHHSPEPDL IHELLGHVPM 360
361 FSDPLLAQMS QDIGLMSLGA SDEHIEKLST VYW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
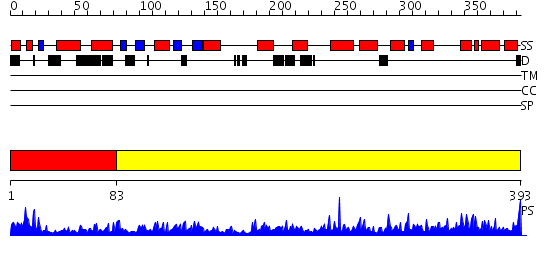
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | 40.045757 | No description for 2qmxA was found. | |
| 2 | View Details | [83..393] | 112.0 | Phenylalanine hydroxylase N-terminal domain; Phenylalanine hydroxylase, PAH |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)