













| General Information: |
|
| Name(s) found: |
acl-7 /
CE31115
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 671 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA |
| Biological Process: |
metabolic process
[IEA phospholipid biosynthetic process [IEA cellular lipid metabolic process [IEA |
| Molecular Function: |
1-acylglycerol-3-phosphate O-acyltransferase activity
[IEA acyltransferase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSASQTGRQ INYPEYENFL EHLKNKGGPF KFVTATKNFP VGSKVGTAPK TRSLAEIRKS 60
61 VLESPRVKSV ISEETLRRKC SPDLVRKEAE EILDEMSHTF NLYNVRSFGY AVCKAMEKLY 120
121 DGIYVNEKKL LEIRERSKKD CVIFMPSHKT YFDFLLLSLI CFQYDIQLPA IAAGQDFMSM 180
181 KFMAGVLRRS GAFFMRRSFG TDQLYWAVFS EYVQTHVVNN DRTVEFFVEA TRSRVGKSLH 240
241 PKYGMLQMVL EPYLRGSVYD IVIVPVSMNY DKILEEKLYA YELLGFPKPK ESTGALLKAR 300
301 EMLKITHGSC FLTFGEPISV RDHFGVSLHR NTFVCQPDSQ FVLDAQAKLE IKRFAHRVVG 360
361 IHNQNAVITV WSVACMAILQ TFDRDDQAII TYCGLYTDVN NLIILQNHLG TVVNIQGNLD 420
421 KNLRYYLTMH ADLFEPFEGS APDFQLKFIA FPVAHQKQDV PIKIMERAVS RLILTTYSNG 480
481 MVHAVDSEGI ICTILRNSGI TDVAKVKEEF CWLHFLMKRE FVTVPGELPQ LFDATLDILR 540
541 ASEILQFTDD NKIEILDEKN AEVLSKLVLP YFYNFEIAFN VFSEKKPMVS SLSEMVGTVQ 600
601 KNISEAYQDR LPNIRLSFLS TEPIKNAFAA LTDFGVFENS NTGMRPNFGK LHQIKNRLQR 660
661 FTQSENHLAK L |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
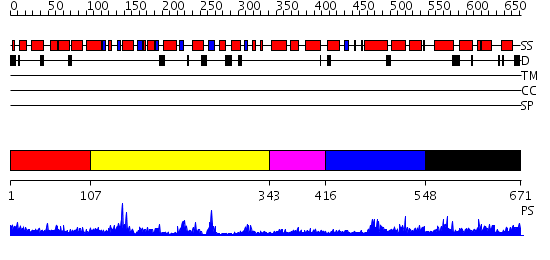
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..106] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [107..342] | 9.0 | The 1.55 A Crystal Structure of Glycerol-3-Phosphate Acyltransferase | |
| 3 | View Details | [343..415] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [416..547] | 1.180988 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [548..671] | 2.094977 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)