













| General Information: |
|
| Name(s) found: |
CE32000
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 705 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
locomotory behavior
[IMP nematode larval development [IMP positive regulation of growth rate [IMP oviposition [IMP mRNA processing [IEA growth [IMP |
| Molecular Function: |
nucleic acid binding
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAFRQGYGVS SYPHYPQQQQ QIVFGQMRMP GQHTPAQQSS QQKPDSSPVF VGNISDKCSD 60
61 EFIRKILNEC GNVASWKRIK GSNGKLQGFG FCHFTDLEGT LRALRILHEF HLGDKKLTVK 120
121 PDEKVRDDLR KNAIENRKRQ GKKELKLKHD ELPADEDDLK KDEEIRLKIL HWMETDHKEL 180
181 FSITEDGELS DETKKVKGHD SKIEKQTVPS SSSNPKKDEH KRNRNRRSRS RTRSRERTSR 240
241 RSRSKESPRS KRSRRSRSSS NSSSESDSSS SRSSSYSSRS RSRTPQRSRT SKRRRSNSHA 300
301 SEDSDDAREK RAFKQMMLDK KQAYLARLKR WESRERQMSK RYEREERKEK DRKKTLQKEG 360
361 KRLKLFLEDY DDEKDDPKYY TSSQFFQRKR DYEREREADQ KDRMQEQQEI EELKRQIMEE 420
421 AANDESINIE EEARKRHKLK EEEAMRKMRA DSGSPNPHQP LGQSANGEKS SSEEESDSEK 480
481 TDVKKEIKDE IKEEPIDVDI SEHVDPNTGS SGTNGNFGWK AIGDDSSLNT KINRPIANGN 540
541 QNQPQIKKEA SPIPIAQRLS GVFGNDDDED DVHSKKKLKP FEITREERMQ VMSAEEKREL 600
601 TKQIIKTIPA TKDELFVHRI EWDQLDGKWM NDRIRPWVAK KVTQFLGEED KSFCDFICDQ 660
661 IEKQATPQEI LKDVAVIIDE DAEQFVIKMW RLLIYEGQAR RLGIT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
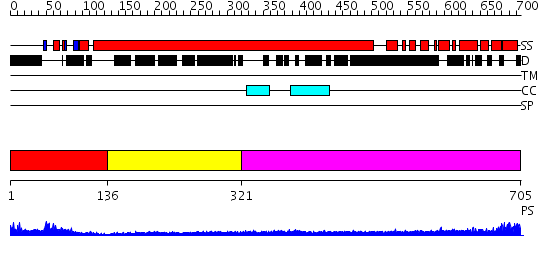
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..135] | 12.39794 | Poly(A)-binding protein | |
| 2 | View Details | [136..320] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [321..705] | 12.154902 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)