













| General Information: |
|
| Name(s) found: |
CE31963
[WormBase]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 758 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
positive regulation of growth rate
[IMP]
receptor-mediated endocytosis [IMP] |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRSVRSSSRT PKPVKRFVFD EDEDLEEIDL DEVDNADFER EDDEEEVLSE PSESPYTSTP 60
61 KSSKRVNKTR GTKDFFKKDE SLQLFRFVLN ECRSQKPGRV GRVAKKRVGV NSMKYWERYK 120
121 REIQGYRLPK QYRYHYSFYS KCFYKVLGLT PEEKVDLYYA CEMPVVTDAE KQWLVEQFHV 180
181 EFDEFGVIIG SDVCTHWDEE HSDSEDDSGK KEEKAREVSR YTEYHDDMMW KFIVDKISEG 240
241 AQPIIDKHPI WDEFVKLNEE NDVISKKSGR NHYDRFRRIL VPNLHFMPYD DFTKAVLYER 300
301 LEYPIPEEYR KILFTTTGAD INESGFIEYL PPSTIHQISP IVPIPCHKSN PKFSQLKEPK 360
361 DLILWSERKK FTPREDSQIW EFVRRKCVDS AGIFVKNDVL RGRGTLFFKD FRNTERKYES 420
421 ITQRFVVLRK LIMDTDYSLK QKLEIYYAIS QPVDEDALDA FKSIACLVLN PDGTIRFAIS 480
481 KSFLIGRISD EPAKLEAENY SYVYDLFIFN ETFGTAENPE PLCSVSQNRA EKFMLVSQLV 540
541 FRFISRFEAL EDRRIAWITP NAQVAPKEPV PAPSKKKSAF EAYQSSPQTF DTIQRFSTKR 600
601 PFFCGDNFQI PPKKPFLAEE DVKLEPMEYE NSEQIVENAE IKLEPAEELL DAETVTVEEF 660
661 LMNSYSTAAP STSTAPAPPK APVTAPPAPQ KSAQLLGKID FAIGKMRENF DRFLQFAQQN 720
721 SADLTVVQRY RYDAQMQQMT EKFTKDVSKV LSGDSIKR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
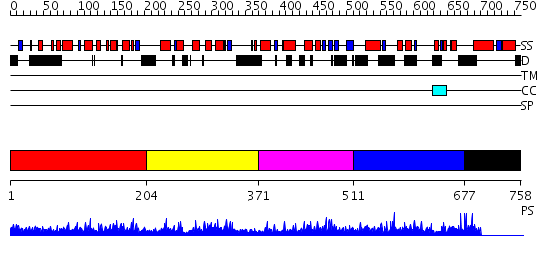
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..203] | 3.02898 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [204..370] | 9.054988 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [371..510] | 8.038995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [511..676] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [677..758] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)