













| General Information: |
|
| Name(s) found: |
cdc-14 /
CE30868
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 693 amino acids |
Gene Ontology: |
|
| Cellular Component: |
spindle
[IDA midbody [IDA |
| Biological Process: |
DNA repair
[IMP positive regulation of growth rate [IMP protein amino acid dephosphorylation [IEA dephosphorylation [IEA |
| Molecular Function: |
nucleic acid binding
[IEA protein tyrosine phosphatase activity [IEA phosphatase activity [IEA protein tyrosine/serine/threonine phosphatase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MREDHSPRRN NTIENTLTEL LPNRLYFGCF PNPDAIDKSD KSVKKTCFIN INNKFHYEPF 60
61 YEDFGPWNLS VLYRLCVQVG KLLEVEEKRS RRVVLFCQDD GTGQYDKIRV NTAYVLGAYL 120
121 IIYQGFSADD AYLKVSSGET VKFVGFRDAS MGSPQYLLHL HDVLRGIEKA LKFGWLDFSD 180
181 FDYEEYEFYE RVENGDFNWI IPGKILSFCG PHNESREENG YPYHAPDVYF DYFRENKVST 240
241 IVRLNAKNYD ASKFTKAGFD HVDLFFIDGS TPSDEIMLKF IKVVDNTKGG VAVHCKAGLG 300
301 RTGTLIACWM MKEYGLTAGE CMGWLRVCRP GSVIGPQQPY LIEKQKFCWS LSQSNGVHLT 360
361 QNKEEKRNVR RLVNQVDDIN LGEERISPKS RENTRPNILR RRVQVQNGRS TAPVTIAPAG 420
421 TSESRRSTKP SRVVDETALD DQGRSQGDRL LQLKAKHQHE SETTSPNSSS SRRFVKSSTP 480
481 QMTVPSQAYL NRNREPIIVT PSKNGTSSGT SSRQLKTTPN GNVAYRTRNS SGNTTSTLTR 540
541 TPASAVFPSM ASRRSETTRY LSPTTPIKPM SPSYTDGTSP RYKARLRSEN PIGSTTSTPF 600
601 SLQPQITKCS LTAESKPPKR ILSMPGTSKS TSSLKKIQVS RPRPYPSTGV RVELCANGKS 660
661 YDIRPRKEAH VIPGAGLAAN TEALLGKRRK TAS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
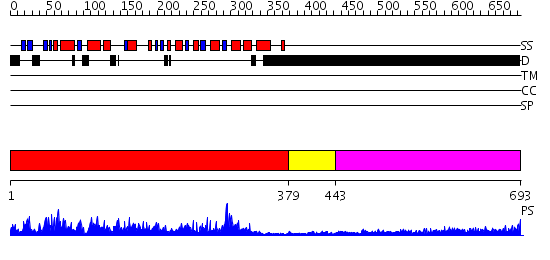
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..378] | 152.0 | Proline directed phosphatase CDC14b2 | |
| 2 | View Details | [379..442] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [443..693] | 4.248995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)