













| General Information: |
|
| Name(s) found: |
ctbp-1 /
CE29966
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 727 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
metabolic process
[IEA L-serine biosynthetic process [IEA |
| Molecular Function: |
nucleic acid binding
[IEA oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor [IEA NAD or NADH binding [IEA cofactor binding [IEA phosphoglycerate dehydrogenase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPTTCGFPNC KFRSRYRGLE DNRHFYRIPK RPLILRQRWL TAIGRTEETV VSQLRICSAH 60
61 FEGGEKKEGD IPVPDPTVDK QIKIELPPKE SKNSDRRRKQ NIPARFPRPE SPSGDSPSYS 120
121 KKSRSFRDFY PSLSSTPSFD PAQSPHTPHP PVLPDPQQAL NDILSMTSTR MNGPSSSRPL 180
181 VALLDGRDCS VEMPILKDVA TVAFCDAQST QEIHEKVLNE AVAALMYHSI KLEKEDLEKF 240
241 KVLKVVFRIG YGIDNIDVKA ATELGIAVCH APGDYVEDVA DSTLSLILDL FRRTYWHAKS 300
301 YSETRKTIGA DQVRENAVGS KKVRGSVLGI LGCGRVGTAV GLRARAFGLH IIFYDPFVRE 360
361 GHDKALGFER VYTMDEFMSR SDCISLHCNL GDETRGIINA DSLRQCKSGV YIVNTSHAGL 420
421 INENDLAAAL KNGHVKGAAL DVHDSVRFDP NCLNPLVGCP NIINTPHSAW MTEASCKDLR 480
481 INAAKEIRKA INGRCPQDLT HCINKEAVMR NSNPINRRTS SAHPLLNMGF PTLPNFPPMS 540
541 MSPHFPYPNP LLAMGAQMGA LNPFMGNGAL PFNPAAALSS LAAAQAANAQ RGSPANRSSR 600
601 SSPSPHTNKS SVSPGNNGHV KTEPSSPAAK IEVDIAENDK HSMMTFLQRL IAPNGDSGAS 660
661 TADSGIEGGD KEKVQSDGDE NMEDMEVIDA EKLKEELNIG QLEEPEEISV GLNNGNRINI 720
721 DEQPLAT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
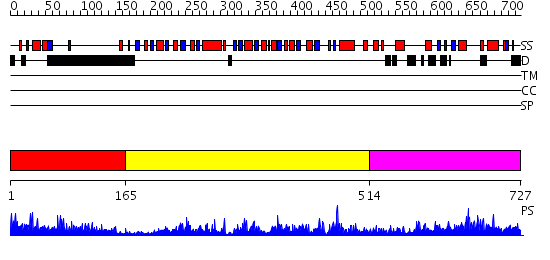
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..164] | 41.69897 | No description for 2jm3A was found. | |
| 2 | View Details | [165..513] | 91.154902 | Transcription corepressor CtbP | |
| 3 | View Details | [514..727] | 8.226994 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)