













| General Information: |
|
| Name(s) found: |
rsr-1 /
CE30346
[WormBase]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 601 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
locomotory behavior
[IMP embryonic development ending in birth or egg hatching [IMP gastrulation with mouth forming first [IMP mRNA processing [IEA |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDAGYFRGAN SEQDGRFSDK EKKLLKTMKF EPQLEQKIDL NRCNMDVIKP WITARVNDIL 60
61 GMEDDVVVEY ILSQIDDKNL NPKLLQINVT GFLNARRARE FVGDLWNLLI EANASEDGIP 120
121 ASLVNQKMAE MKTNDRDEKG DDREDNDWTN RYKTLSNGRY TGPAREKAVR DDRIPARALS 180
181 PRKTPPRRNA SPGGDGGGGS PAARRGGSAG NRRRSPPRRG SPRRGSPRRD TRRSPPRRRG 240
241 SPPARGGDRR DRREDRRPVR DEKEREEESR RREIERKKRH EEAELRQKAI ASVAARAKSG 300
301 SPRRRRSPSA SKSPPPARRR RSPSQSKSPA PKRAKSRSKS PPAPARRRRS PSASKSPPPA 360
361 PKRAKSRSKS PPARRRRSPS ASKSPPAPRR RRSPSKSRSP APKREIPPAR RRRSPSASKS 420
421 PPAPKRAKSR SKSPPAPRRR RSPSQSKSPA PRRRRSPSKS PQAPRRRRSP SGSKSRSPRR 480
481 RRSPAAAPRR RQSPQRRRSP RRRRSPSSSS RSRSPPPPPR RPRQDSEQQA PVAVKSPEMK 540
541 KRRPNDTDSD VSVDSEAEEE RRRKKAKKEK KAAKKHKKEK KSKKHRKRRS PSASESGSES 600
601 D |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
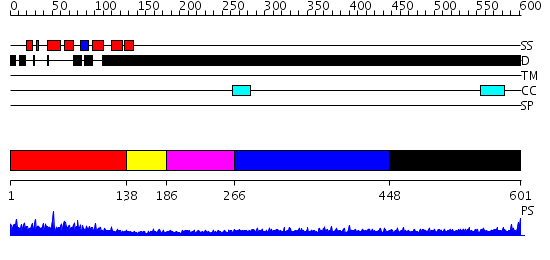
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..137] | 48.045757 | Solution structure of the PWI motif from SRm160 | |
| 2 | View Details | [138..185] | 1.05 | 30S subunit | |
| 3 | View Details | [186..265] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [266..447] | 3.190997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [448..601] | 3.180982 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)