













| General Information: |
|
| Name(s) found: |
duo-2 /
CE40673
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 551 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
ubiquitin-dependent protein catabolic process
[IEA |
| Molecular Function: |
ubiquitin thiolesterase activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGLQWNNVFH QRQDFVHSCT HRVERLLELE ELRKRLVNTG NSCYYNSTLQ AMSSCGPMVV 60
61 RCENLYRMVR YKIRFLDNSL NINQTQPCDI KNNNIVLRNF LKIVSCLQLG GHLNEPNEVM 120
121 KLSTSKLLQY RRNVGKIHEG FNNDDQHDAH EFFLTLIGAV DDVMKVVLEN AVMIDDFESP 180
181 LNPAKVFKID VETLCLVVTL KRYELEGRGS RFSMKNKNCR VEPTFELDAT SLRNFMPVEC 240
241 SELNKNDAHS NNSRLHIGAD EEVVQEDVTA RKQLFFDVFS NMADFDNTLV GMGFANGVNQ 300
301 KRCHFKRERV AHISEMSQYQ LPGVTRKIEA DGNCFYRAVS WWLTGVESHH MIFREAVGKH 360
361 LKKNEPKFKK YCHDEIYEKY VDNVMREGVW ASTCEIFAMA NMLNVAIMTY LGDSGWMPHS 420
421 PQNTCPPRQG ALYLKNTHMH YESTISLKKN TSMKSTMTSQ KGNNNERLMH GELHEKDNMR 480
481 GDAPPRRYTL VSVVCHLGSS PNKGHHVAYT KKSLDNDGWL RCSDDKLRDV SKENVANAVR 540
541 SSGYVLYYIL Q |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
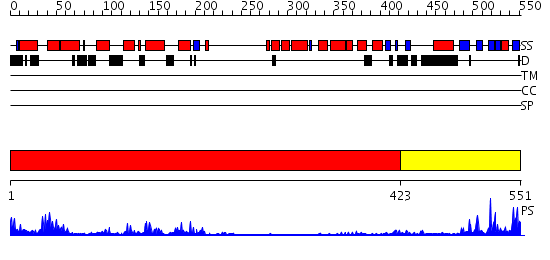
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..422] | 23.0 | Crystal structure of HAUSP | |
| 2 | View Details | [423..551] | 6.06 | Crystal structure of HAUSP |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)