













| General Information: |
|
| Name(s) found: |
ptb-1 /
CE30508
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 615 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
mRNA processing
[IEA |
| Molecular Function: |
nucleic acid binding
[IEA RNA binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTKRGPDDLQ LSLRNGVGGA STTAPGVAAA AYYANVMTAD SEPKKAKLDP TLYSSQFYTP 60
61 AMHHGLMTAD ASTVAAVTAA ANATTAQLLT GNPITQIQGI QLSTLPQCNS TSAVTSIPVS 120
121 KVVHVRNIPP DLVDVELMQL CIQYGPVSNY MMLKGKSQAF VEYEEEASAA AFVSGMTAVP 180
181 IQIRGRTLFA QYSTHRELKF DKNKAISDTE SVANGSVSNF EVGTQQQPNS VLRTIIENMM 240
241 FPVSLDVLYQ LFTRYGKVLR IITFNKNNTF QALVQMSEAN SAQLAKQGLE NQNVYNGCCT 300
301 LRIDYSKLST LNVKYNNDKS RDYTNPNLPA GEMTLEQTIA MSIPGLQNLI PANPYNFAFG 360
361 ANPATTFLTT QLAASTAAAA AVNDSANAAA LAPYLNPLGL TSANLAPSIS SMRFPMINLT 420
421 PVILVSNLHE MKVTTDALFT LFGVYGDVMR VKILYNKKDN ALIQYSEPQQ AQLALTHLDK 480
481 VKWHDRLIRV APSKHTNVQM PKEGQPDAGL TRDYAHSTLH RFKKPGSKNY LNIYPPCATL 540
541 HLSNIPTSVS EEKLKEMFAE AGFAVKAFKF FPKDHKMALC QLEDIETAID ALIAMHNHKL 600
601 AENAHLRVSF SKSGI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
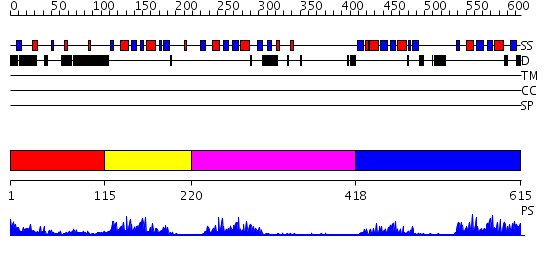
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..114] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [115..219] | 16.09691 | solution structure of RNA binding domain in PTB-like protein L | |
| 3 | View Details | [220..417] | 17.0 | Solution structure of Polypyrimidine Tract Binding protein RBD2 complexed with CUCUCU RNA | |
| 4 | View Details | [418..615] | 41.0 | Solution structure of Polypyrimidine Tract Binding protein RBD34 complexed with CUCUCU RNA |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.55 |
Source: Reynolds et al. (2008)