













| General Information: |
|
| Name(s) found: |
lag-1 /
CE28839
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 673 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][IEA |
| Biological Process: |
positive regulation of vulval development
[IMP collagen and cuticulin-based cuticle attachment to epithelium [IMP molting cycle, protein-based cuticle [IMP regulation of cell fate specification [IMP hermaphrodite genitalia development [IMP nematode larval development [IMP embryonic development ending in birth or egg hatching [IMP oviposition [IMP germ-line stem cell division [IMP positive regulation of transcription from RNA polymerase II promoter [IMP cell fate specification [IMP |
| Molecular Function: |
DNA binding
[IEA transcription factor activity [IEA protein binding [IEA sequence-specific DNA binding [IDA] Notch binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPLAYSTHDN FYESPKTPQP TWDHVHAQFP LGEPARNLDK FIVPEAMFQS VSPLAGVAAA 60
61 PSQIAALQQI QALMTFQMQQ NNLFPKIDTI SKSPTPELAS PSAKRMRLSP STSSHSDVAS 120
121 TSKGTNGQDQ SKNSPTNSDP FYTIGQSTPN TSSFLDTSNS SFGAPSTVVA NPMTNYQLAF 180
181 QAKLGSLHSL IGDSVQSLTS DRMIDFLSNK EKYECVISIF HAKVAQKSYG NEKRFFCPPP 240
241 CIYLIGQGWK LKKDRVAQLY KTLKASAQKD AAIENDPIHE QQATELVAYI GIGSDTSERQ 300
301 QLDFSTGKVR HPGDQRQDPN IYDYCAAKTL YISDSDKRKY FDLNAQFFYG CGMEIGGFVS 360
361 QRIKVISKPS KKKQSMKNTD CKYLCIASGT KVALFNRLRS QTVSTRYLHV EGNAFHASST 420
421 KWGAFTIHLF DDERGLQETD NFAVRDGFVY YGSVVKLVDS VTGIALPRLR IRKVDKQQVI 480
481 LDASCSEEPV SQLHKCAFQM IDNELVYLCL SHDKIIQHQA TAINEHRHQI NDGAAWTIIS 540
541 TDKAEYRFFE AMGQVANPIS PCPVVGSLEV DGHGEASRVE LHGRDFKPNL KVWFGATPVE 600
601 TTFRSEESLH CSIPPVSQVR NEQTHWMFTN RTTGDVEVPI SLVRDDGVVY SSGLTFSYKS 660
661 LERHGPCRIV SNY |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
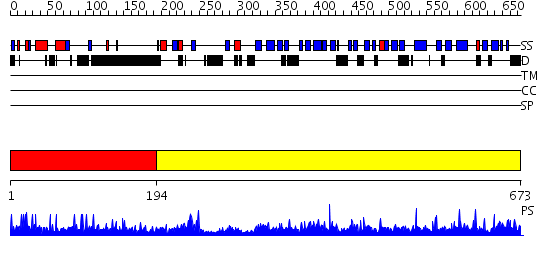
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..193] | 1.02297 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [194..673] | 1000.0 | Crystal Structure of CSL bound to DNA |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)