













| General Information: |
|
| Name(s) found: |
gap-2 /
CE28277
[WormBase]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 787 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA |
| Biological Process: |
regulation of small GTPase mediated signal transduction
[IEA reproduction [IMP |
| Molecular Function: |
GTPase activator activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRGEYDPWDP SFHNSSMIPY SLASIYPSIE NLPEEFSNEN NKIKNVLKNF IWSFKLKKNY 60
61 VRVSSLFGEY TVNTSHSDSG TSRIASALGG KSSSQESPSL RIKARWQSVH ILPLRAYDNL 120
121 LETLCYNYLP LCEQLEPVLN VRDKEDLATS LVRVMYKHNL AKEFLCDLIM KEVEKLDNDH 180
181 LMFRGNTLAT KAMESFMKLV ADDYLDSTLS DFIKTVLQCE DSCEVDPQKL GNVSNSSLEK 240
241 NRALLMRYVE VAWTKILNNV HQLPKNLRDV FSALRCRLEA QNREALADTL ISSSIFLRFL 300
301 CPAILSPSLF NLVSEYPSPT NARNLTLIAK TLQNLANFSK FGGKEPHMEF MNEFVDREWH 360
361 RMKDFLLRIS SESKSGPEKN ADAIVDAGKE LSLIATYLEE AWTPLLQEKN GNKHPLSNVK 420
421 SVLSELAECK RRSDNGVFHS PMVQQPSSDY ENSPQQHVVP RHENVPAYRS TPPTGQATVL 480
481 GRSTNRPATH LLTSDDYVLS SAFQTPSLRP GGTRLSDETG TSSSRTSDKT TSSAEIRDDT 540
541 DSDFELREDR GRGGRNRKRL PRTDASPSSS QQASSGYLSN NPSRSSYSNS SSSSPVERMA 600
601 ALSIANPVFG PGPSSGYAIP AEPKEIVYQK RASPPPYDPD VHNYHYQPMQ VYAVPPDCQV 660
661 SPRTQATGGV NAQNRLSLPR TNPRASRNST LLRPSVVNVP DDWDRTSDYW RDRGENNYRS 720
721 QLESQVESQA REIERLMREN IELKSKMMSS TKTVDSKRSD SGASEDSYDS LSSLDRPSRQ 780
781 SLVVVPN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
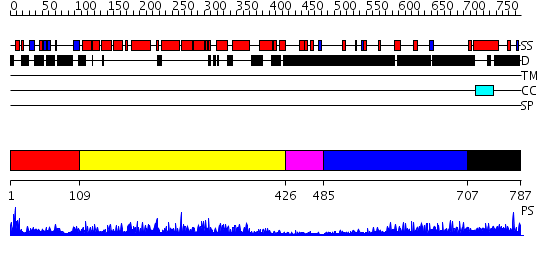
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..108] | 5.0 | No description for 2nsqA was found. | |
| 2 | View Details | [109..425] | 76.30103 | RAS-GTPASE-ACTIVATING DOMAIN OF HUMAN P120GAP | |
| 3 | View Details | [426..484] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [485..706] | 3.69897 | No description for 1y0fB was found. | |
| 5 | View Details | [707..787] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)