













| General Information: |
|
| Name(s) found: |
unc-43 /
CE28058
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 571 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
protein amino acid phosphorylation
[IEA |
| Molecular Function: |
magnesium ion binding
[IEA ATP binding [IEA protein tyrosine kinase activity [IEA protein kinase activity [IEA protein serine/threonine kinase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMNASTKFSD NYDVKEELGK GAFSVVRRCV HKTTGLEFAA KIINTKKLSA RDFQKLEREA 60
61 RICRKLQHPN IVRLHDSIQE ESFHYLVFDL VTGGELFEDI VAREFYSEAD ASHCIQQILE 120
121 SIAYCHSNGI VHRDLKPENL LLASKAKGAA VKLADFGLAI EVNDSEAWHG FAGTPGYLSP 180
181 EVLKKDPYSK PVDIWACGVI LYILLVGYPP FWDEDQHRLY AQIKAGAYDY PSPEWDTVTP 240
241 EAKSLIDSML TVNPKKRITA DQALKVPWIC NRERVASAIH RQDTVDCLKK FNARRKLKAA 300
301 ISAVKMVTRM SGVLRTSDST GSVASNGSTT HDASQVAGTS SQPTSPAAEG AILTTMIATR 360
361 NLSNLGRNLL NKKEQGPPST IKESSESSQT IDDNDSEKGG GQLKHENTVV RADGATGIVS 420
421 SSNSSTASKS SSTNLSAQKQ DIVRVTQTLL DAISCKDFET YTRLCDTSMT CFEPEALGNL 480
481 IEGIEFHRFY FDGNRKNQVH TTMLNPNVHI IGEDAACVAY VKLTQFLDRN GEAHTRQSQE 540
541 SRVWSKKQGR WVCVHVHRST QPSTNTTVSE F |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | hcp2 ip from december | Cheeseman, I.M., et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
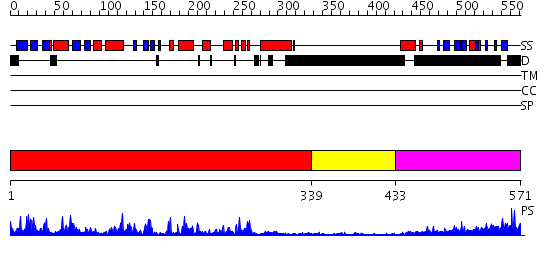
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..338] | 93.221849 | Crystal Structure of the Auto-Inhibited Kinase Domain of Calcium/Calmodulin Activated Kinase II | |
| 2 | View Details | [339..432] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [433..571] | 39.522879 | The Association Domain of C. elegans CaMKII |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.78 |
Source: Reynolds et al. (2008)