













| General Information: |
|
| Name(s) found: |
vhp-1 /
CE27919
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 606 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS cytoplasm [ISS |
| Biological Process: |
inactivation of MAPK activity
[IDA nematode larval development [IMP morphogenesis of an epithelium [IMP protein amino acid dephosphorylation [IDA response to copper ion [IGI negative regulation of JNK cascade [IDA dephosphorylation [IEA positive regulation of locomotion [IMP |
| Molecular Function: |
protein tyrosine phosphatase activity
[IEA phosphatase activity [IEA JUN kinase phosphatase activity [IDA MAP kinase tyrosine/serine/threonine phosphatase activity [IDA protein tyrosine/serine/threonine phosphatase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGGQPFEAFR KNRKRKKTKN KKKRRNNNNS KNKTPNTFPN EIEEQDPVSS LPTFPAKKFG 60
61 LKLQLTLTSS PTNSSSPISS SSPTNGGFKQ FAQQYPQLCE SSEGMTRLPQ SLSQPCLSQP 120
121 TGDGITLITP NIYLGSQIDS LDETMLDALD ISVVINLSMT CPKSVCIKED KNFMRIPVND 180
181 SYQEKLSPYF PMAYEFLEKC RRAGKKCLIH CLAGISRSPT LAISYIMRYM KMGSDDAYRY 240
241 VKERRPSISP NFNFMGQLLE YENVLIKDHV LDYNQASRPH RHMDYYGPSD LCPPKVPKSA 300
301 SSNCVFPGST HDESSPSSPS VSEGSAASEP ETSSSAASSS STASAPPSMP STSEQGTSSG 360
361 TVNVNGKRNM TMDLGLPHRP KALGLPSRIG TSVAELPSPS TELSRLSFNG PEAIAPSTPI 420
421 LNFTNPCFNS PIIPVASSSR EVILTLPTPA ASSSSSTSSE PSFDFSSFES SSSSSIVVEN 480
481 PFFASTEVPA GSSSISTPSG SQSTPASASS SAASRCRMKG FFKVFSKKAP ASTSTPASST 540
541 PGTSRAARPE CLRSSGIIIS APVLAITEEE DAESPESGFN EPEVGEEDDD SVSICSTSSL 600
601 EIPCHQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
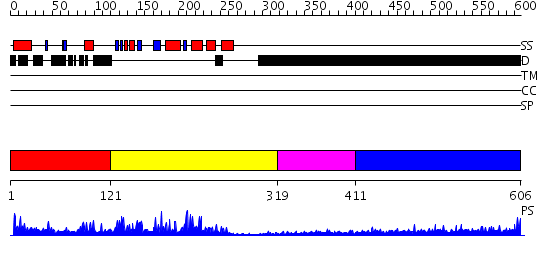
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..120] | 22.69897 | No description for 2oucA was found. | |
| 2 | View Details | [121..318] | 42.0 | No description for 2g6zA was found. | |
| 3 | View Details | [319..410] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [411..606] | 11.198995 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)