













| General Information: |
|
| Name(s) found: |
vhp-1 /
CE27918
[WormBase]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 657 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS cytoplasm [ISS |
| Biological Process: |
inactivation of MAPK activity
[IDA nematode larval development [IMP morphogenesis of an epithelium [IMP protein amino acid dephosphorylation [IDA response to copper ion [IGI negative regulation of JNK cascade [IDA dephosphorylation [IEA positive regulation of locomotion [IMP |
| Molecular Function: |
protein tyrosine phosphatase activity
[IEA phosphatase activity [IEA JUN kinase phosphatase activity [IDA MAP kinase tyrosine/serine/threonine phosphatase activity [IDA protein tyrosine/serine/threonine phosphatase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTVLDTVTIS TCGLAALIRE APDTTLVVDC RGFTEYNESH VRHSMNAFFS KLIRRRLFEN 60
61 KLDDNCLIHQ LMSCSSGCTK MDEKLDLVLY AEEDKPRGNK RRIASCNAPE STAKIMRVLR 120
121 ERLEDTDKFR SVMVLEGGFK QFAQQYPQLC ESSEGMTRLP QSLSQPCLSQ PTGDGITLIT 180
181 PNIYLGSQID SLDETMLDAL DISVVINLSM TCPKSVCIKE DKNFMRIPVN DSYQEKLSPY 240
241 FPMAYEFLEK CRRAGKKCLI HCLAGISRSP TLAISYIMRY MKMGSDDAYR YVKERRPSIS 300
301 PNFNFMGQLL EYENVLIKDH VLDYNQASRP HRHMDYYGPS DLCPPKVPKS ASSNCVFPGS 360
361 THDESSPSSP SVSEGSAASE PETSSSAASS SSTASAPPSM PSTSEQGTSS GTVNVNGKRN 420
421 MTMDLGLPHR PKALGLPSRI GTSVAELPSP STELSRLSFN GPEAIAPSTP ILNFTNPCFN 480
481 SPIIPVASSS REVILTLPTP AASSSSSTSS EPSFDFSSFE SSSSSSIVVE NPFFASTEVP 540
541 AGSSSISTPS GSQSTPASAS SSAASRCRMK GFFKVFSKKA PASTSTPASS TPGTSRAARP 600
601 ECLRSSGIII SAPVLAITEE EDAESPESGF NEPEVGEEDD DSVSICSTSS LEIPCHQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
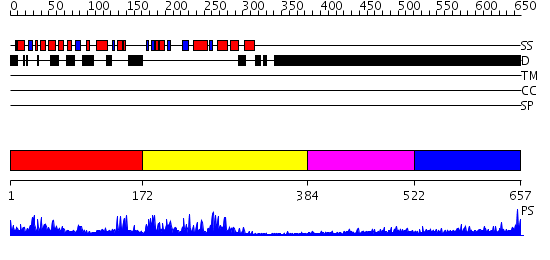
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..171] | 32.0 | Erk2 binding domain of Mapk phosphatase mkp-3 | |
| 2 | View Details | [172..383] | 41.39794 | No description for 2g6zA was found. | |
| 3 | View Details | [384..521] | 1.227996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [522..657] | 4.217995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)