













| General Information: |
|
| Name(s) found: |
dnj-25 /
CE27221
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 784 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
molting cycle, protein-based cuticle
[IMP DNA repair [IMP nematode larval development [IMP positive regulation of growth rate [IMP receptor-mediated endocytosis [IMP growth [IMP |
| Molecular Function: |
heat shock protein binding
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVFDEEPPAQ KKWTQQATTS AFGLMESLKG QGSSLAKNLK NFGKSLVKEH KSQGATTPPE 60
61 HRDDDIRLTW LTNRIVLADT LHKSKEGLLK AETAVVREMI DCQPYVVVNV HAEPLKLDGY 120
121 AKSVHVPLGS AAKGGLPKYP TIQALVPILN EYCLLTNDAA IVIFGNDENV QITAAFCLIA 180
181 SRTISRVSEF MSDSLPNRFD RLPVTYHLFL EQTKHVCKYH ARRQQLQSGA ILNQIILEPG 240
241 TLLIDGDLYV AIQQGINQLK VVEFSANSCR NRQGQLVFNM ESTETVDDVV IFLGKKSDVK 300
301 HPILASKFNT QLLHSEDSSI IFTSSEMDVS KMIASAMPVQ DLRMIVNFAT RRMPTLNTFQ 360
361 FDRYKLLCVR NDRELDGYQR SYGDVDSEDE SPILRSARSK SGTKAAETSS SQGNFFDSLQ 420
421 FEDRQEPHHF REQRDSTLLD HHDDVTHHTA DLLDGFTIGG TEKLSPANET PILSAPTPAS 480
481 SDYDSMTSSQ NRRNPSPATA IDDLLGLGTE TSAPAPAPST HFNNSSSGAN LVDFDLGAPI 540
541 KPTSSASNLS TASPATSSWS NTDLLGGFSP MKPQTTSTPP NTRNPSASNS KPVSQVNSQA 600
601 DFEAFLSNYP EQRQAAPQNT QNVSQKTQQT RPQNTYKPAF GDAATGSAKV SMDAFSDLLN 660
661 QGGFKPTQRE NQKQSIGNML KVEQSKNLTP EEIQIRDWTQ GKERNIRALL GSLHNVLWEG 720
721 ADRWNQPSMG DLLTPDQIKK HYRKACLVVH PDKLTGSPHL SLAKMAFTEL NDAYSKYQND 780
781 PAAM |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | hcp1 IP from december2004 | Cheeseman, I.M., et al. (2005) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
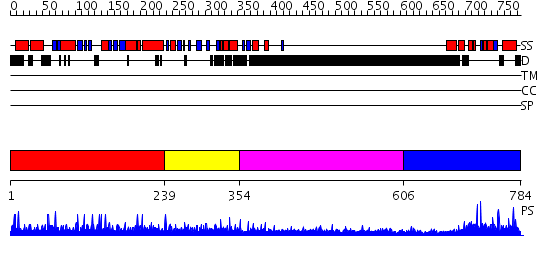
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..238] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [239..353] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [354..605] | 4.270996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [606..784] | 57.09691 | NMR Structure of the J-Domain and Clathrin Substrate Binding Domain of Bovine Auxilin |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)