













| General Information: |
|
| Name(s) found: |
xnd-1 /
CE26863
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 702 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
reproduction
[IMP |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSEPIVANL DNSMTTEPAP AAFPPISPMS FAFMNEEERA AVKFPKVDPK PQTAKDDEPS 60
61 TSQKTLSVDD LLIVDDDDET DSPPASSSNY EPKAHIGWAS FGDSCLPPPP KPVKKATDPN 120
121 LPRRKVYVIR SQNTDKSKSP LVVNNQQVTL EKAQNSPKNP NPVVSKPIVL TDSDEDVDVV 180
181 GFDEEEEAIK IMADPTRPPD LPPQRSLISR FESNRTKRDI FRNSYDSDEE EFLRSRYQRA 240
241 VTPPPILERQ SGSTRESVSE PSEGKILEED ATLGSEHTER VGKRIELEEI NPSQLLRTKI 300
301 SASAIPILPL SKSIMERKKV ALEMTKNAVI RQANNFRPKA KNSTVAPKSV QIPAYNHKTP 360
361 MTFYSNMAAP AFVKGVNGRC YRCAEIQKPV DMSSFRFVDD STVIAIRAHL CDQTRVMVAR 420
421 TAIWNREYAK RLGDRAAGTV WQKPDEVSAQ MTGFCSATVR RCIEIANMSI IPKCADRVGV 480
481 SRNELTSLKS SFGSEKFCGQ TMRAPHVLTQ FYSVNQASMA RNREGSDGPS SSAARPVGRP 540
541 PTTQPVETAV EKKKNDEDEK RHHPLTNFTT ASTSSQNLQD QPPPKNTVLR WSNITTPRIL 600
601 RPPTLKTPYS LPRTAVIPVM KKKQPEIEQV QPKKDTTASL VRKRGRPRKE KPLEVQSPRK 660
661 GLYLRFKSCT KYIVRTPIDK SVQETVNYLL DEVDKRASTS AT |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | hcp1 IP from december2004 | Cheeseman, I.M., et al. (2005) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
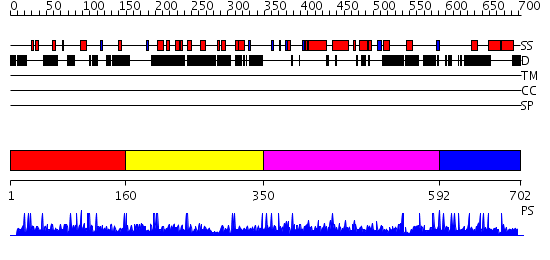
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..159] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [160..349] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [350..591] | 1.004945 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [592..702] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)