













| General Information: |
|
| Name(s) found: |
psa-1 /
CE25208
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 789 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
negative regulation of vulval development
[IMP hermaphrodite genitalia development [IMP regulation of oviposition [IMP nematode larval development [IGI morphogenesis of an epithelium [IMP embryonic development ending in birth or egg hatching [IGI positive regulation of growth rate [IMP gonad morphogenesis [IMP positive regulation of multicellular organism growth [IGI reproduction [IGI growth [IMP |
| Molecular Function: |
DNA binding
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKKGASKRRR DDDDGDVSMT GDDDDSRSGA AVKVEVPKGK EKDAEFSAPK GQKLTDLDEE 60
61 GVQSTKEAPQ LAEGNVIEQT HYIVVPSYAG WFDYNAIHQI EKRAMPEFFN GKNKSKTPDV 120
121 YVAYRNFMVD TYRLNPFEYV SATACRRNLA GDVCSIVRLH SFLEQWGLLN YQVDSDARPA 180
181 PVAPPPTSHF MVLADTPTGI QPMNPPGKES AGASGEPPKE EIKTEIESIS TPGLKIDQYQ 240
241 KQAIAMRTKG APPGRDWTEQ ETCLLLEALE MFKDDWNKVC DHVGTRTQHE CVLKFLQLPI 300
301 QDPYLTENLS SDKAEAAPGG AAKEVLGPLA FQPVPFSQSG NPVMSTVAFL ASVVDPQVAA 360
361 AATKAAMEEF GKLKEEIPPL VAEAHEKNVA AMAEKTGQVD GAVGLTKSGL KPAEEAAGDS 420
421 DEKMDTNTND DVPSTTEAKS AIDKGVQAAA ASCLAAAAVK AKHLAQIEER RIKSLVAQLV 480
481 ETQMKKLEMK LRHFDELEQI MDKERESLEY QRHQLILERQ AFHMDQLKYL ENRAKHEAHS 540
541 RMTSSGALPA GLPPGFEVTG PPQPTPQVQI SAQEAAIPEK MDTSEAATAA RPPSTPQAPQ 600
601 APPVQAAPAP VQAPQAPQAP PQAYQGYGGP GGPPQQAYRY PPQQGQQYSP YPPPQQQQQH 660
661 QAQQAQSQAH YGPPGGGQGP PPPPQGQQYY GGPPPPGQPY GPPGGYPPQQ QRPPYQAQPY 720
721 PGPPPPQQQR GYGYPPPPQP GHPYQQPYGQ MPPPPHGQYQ PQQQQGGPMG PPGGHHEGGD 780
781 GGDSQQKQE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | hcp1 IP from december2004 | Cheeseman, I.M., et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
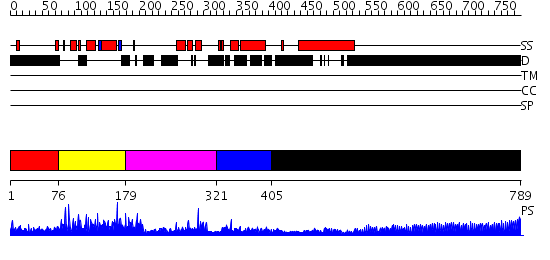
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..75] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [76..178] | 44.39794 | Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes | |
| 3 | View Details | [179..320] | 2.7 | No description for 2nogA was found. | |
| 4 | View Details | [321..404] | 2.9 | CHICKEN B-MYB DNA BINDING DOMAIN, REPEAT 2 AND REPEAT3, NMR, 32 STRUCTURES | |
| 5 | View Details | [405..789] | 28.09691 | No description for 1y0fB was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)