













| General Information: |
|
| Name(s) found: |
gap-2 /
CE23476
[WormBase]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 790 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA |
| Biological Process: |
regulation of small GTPase mediated signal transduction
[IEA reproduction [IMP |
| Molecular Function: |
GTPase activator activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRGEYDPWDP SFHNSSMIPY SLASIYPSIE NLPEEFSNEN NKIKNVLKNF IWSFKLKKNY 60
61 VRVSSLFGEY CRYTVNTSHS DSGTSRIASA LGGKSSSQES PSLRIKARWQ SVHILPLRAY 120
121 DNLLETLCYN YLPLCEQLEP VLNVRDKEDL ATSLVRVMYK HNLAKEFLCD LIMKEVEKLD 180
181 NDHLMFRGNT LATKAMESFM KLVADDYLDS TLSDFIKTVL QCEDSCEVDP QKLGNVSNSS 240
241 LEKNRALLMR YVEVAWTKIL NNVHQLPKNL RDVFSALRCR LEAQNREALA DTLISSSIFL 300
301 RFLCPAILSP SLFNLVSEYP SPTNARNLTL IAKTLQNLAN FSKFGGKEPH MEFMNEFVDR 360
361 EWHRMKDFLL RISSESKSGP EKNADAIVDA GKELSLIATY LEEAWTPLLQ EKNGNKHPLS 420
421 NVKSVLSELA ECKRRSDNGV FHSPMVQQPS SDYENSPQQH VVPRHENVPA YRSTPPTGQA 480
481 TVLGRSTNRP ATHLLTSDDY VLSSAFQTPS LRPGGTRLSD ETGTSSSRTS DKTTSSAEIR 540
541 DDTDSDFELR EDRGRGGRNR KRLPRTDASP SSSQQASSGY LSNNPSRSSY SNSSSSSPVE 600
601 RMAALSIANP VFGPGPSSGY AIPAEPKEIV YQKRASPPPY DPDVHNYHYQ PMQVYAVPPD 660
661 CQVSPRTQAT GGVNAQNRLS LPRTNPRASR NSTLLRPSVV NVPDDWDRTS DYWRDRGENN 720
721 YRSQLESQVE SQAREIERLM RENIELKSKM MSSTKTVDSK RSDSGASEDS YDSLSSLDRP 780
781 SRQSLVVVPN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
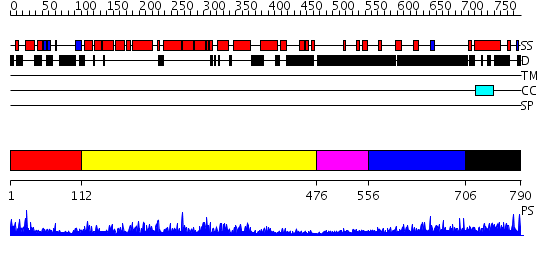
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..111] | 4.221849 | Synaptogamin I | |
| 2 | View Details | [112..475] | 74.0 | RAS-GTPASE-ACTIVATING DOMAIN OF HUMAN P120GAP | |
| 3 | View Details | [476..555] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [556..705] | 7.228997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [706..790] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)