













| General Information: |
|
| Name(s) found: |
CE20170
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 783 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
nematode larval development
[IMP]
locomotion [IMP] growth [IMP] reproduction [IMP] |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIIYTTPTAL ILEEQGERVE FDRRNGKVLE KPEESLEILD QDVVLCGNVE FLIGKISFDE 60
61 QHFLFFVVDS SVVACYRGTS AESSHSIRRI ERVIAINVKS DGSVIKSQIT PGSGTKLKQG 120
121 TEKIMKFFAE KVTKAEPRPA LLDDVLKLFN DSKDFYFCRS RDVTISSQKY FEKREAHTSE 180
181 DSFFWNKRMV GNLGEAKISD KFTCPIMQGY VATSQLEITD QINAYLTITI ISRRSTRRAG 240
241 ARYLRRGIDE ASNVANFVET ELILNIFEHE LSFVQCRGSI PVFWSQRGFK YRPPLIINRS 300
301 VEETHGVFTE HFKRLKAHYD TPLVAVSLVD QRGRELPLAQ RFLEHCVKAD DNDVTFFSFD 360
361 LHQHCRGLNF QKLQTLISSM EETLKTIGFC WVDKTGEVVQ SQKGVVRTNC IDCLDRTNLV 420
421 QGQISLFVVL QQAQRLGIFG PLCEPPEVLV QTMQTMWADN GDVISTQYAG TAALKGDVTR 480
481 NGERKLMGVM KDGYNSASRY YLTHTKDAQR QKAINIVTGQ TEADENAEEE ENEKEEEENI 540
541 SRMVTETIQF LVPTQQTVIA GWGLINAFQS SDEIDTVVIL TRSNLYIVTY EIDGEKMTDV 600
601 QIVALEDINT IQTGTSGNNR QCARLETLEG VFIWRPSTVR LFNNTAIRLK SLEEANEYIS 660
661 SVAEQIQVGR NMLTGAEGSV VPVAKITIPQ MSVHRKSMAA MGALFGKIKR VGKPSSLSKS 720
721 ASTISTAQLP ETDGNESDAS QKSDHLMTKI LKMSQFKSDS PQPEPFSSLL PAISECQTKI 780
781 SLL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
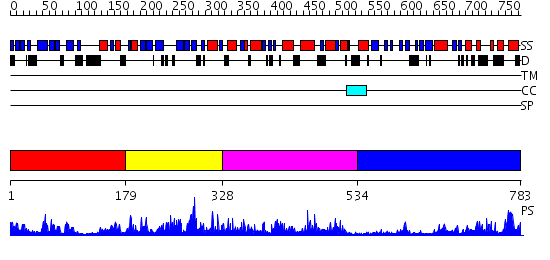
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..178] | 7.172989 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [179..327] | 1.175974 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [328..533] | 2.223986 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [534..783] | 41.0 | Synaptojanin, IPP5C domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)