













| General Information: |
|
| Name(s) found: |
ape-1 /
CE19874
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 769 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
cell death
[IMP |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVTTSSGGGI GYPANNGVTQ VSLIHSSDSV RTVSTAPIYR PTSSMASTMA HKSSTAPFIS 60
61 ANQRMSKPPV RVVAQPPPPH PQALSQQYHQ QNPMMMYSAP NTRPHVIPTM QVQPTMAAQI 120
121 KRNNPVNAQF QNPSEMIADY GVKPQSVEMV QRVRAVRRQV ADEETELRRL RELEHETAQL 180
181 QNKNYGRERE LNVQGSMLKE AQLELRNASM RAQSLNKHLE EMYRRRQTAA AAALVEQRKM 240
241 QQHQILLARA ANQVSTQEVI RPRASVEPFQ VNNTQQQQPS PQMMKSEEFS EKRDLNGQTG 300
301 SYDAIDGSGD HQKIPTEPSY LAPCKENQQK YSELSKMAST DPHSNHSSPS TSSQKAPTLI 360
361 TFSPPSFEQK INSSTMTRDS PFVERPTSFG DSLDESRLRS GKTDLVSLRS DSLKATKRRS 420
421 WAASEGTSMS EAEMIHRLLD EQRRGRSHFI PQLPTSQEEP SAITSETYAE EVVNSESKQV 480
481 ATSSDSTNNL ELPTEQMVLG SDTTTEEDAS SCSTRSDDGQ NLEMEVAIER RTVKGILRRP 540
541 NEKMNKGRIE FDPLALLLDA ALEGELDLVR SSASKLTDVS QANDEGITAL HNAICAGHYE 600
601 IVRFLIENDA DVNAQDSDGW TPLHCAASCN NLPMVRQLVE GGGCVLASTL SDMETPVEKC 660
661 EEDEDGYDGC LKYLSAAHNS TGSINTGKVY AAYGYEAAFE DELSFDAGDE LTVIEKDKVD 720
721 KNWWTCEKNN GEKGQVPRTY LALYPSLKYR KKLNFVMFDL PLESNNNVE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
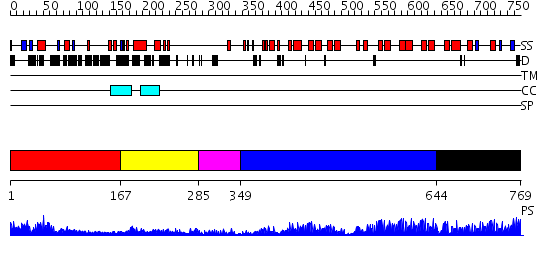
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..166] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [167..284] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [285..348] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [349..643] | 27.69897 | Transcription factor inhibitor I-kappa-B-beta, IKBB | |
| 5 | View Details | [644..769] | 32.39794 | 53BP2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 |
|
|||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)