













| General Information: |
|
| Name(s) found: |
CE18521
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 703 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
nitrogen compound metabolic process
[IEA NAD biosynthetic process [IEA |
| Molecular Function: |
hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds
[IEA NAD+ synthase (glutamine-hydrolyzing) activity [IEA ATP binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNQWDRRCRV ATCTVNNWAL DFKGNYERIV KTCEEAAALG ARIRLGPELE IPGYGCADHF 60
61 FELDTERHSW EMLSKLVEKS KKWPNLLVVT GLPTRFRGLL YNCAAALRNG KLLFIRAKMG 120
121 LADDNVYRES RWFVKWTETF KHYQMPLNSD IHFDQETVPF GDGILESSDN VRIGFEICEE 180
181 LWSARSTNVR LAEQGVDIMC NGSGSHHILG KSNYRINQLI LGSSAKVGGV YLYANQRGCD 240
241 GDRVYYDGAS SVAQNGDLLA QIHQFDIEDT SVVSAVVDLS DNQCFRHMKS SDRGNASDQV 300
301 TVVPIRFDGK MTGGIKYNEK STAPIHNVED LQLSPIAELC HGPPAYLWTY LRRSGMAGYF 360
361 IPLSGGQDSS AVAAMVRLMC EKVCGAIKRR RETDGGDDPA YYLGGKKVGE DPAELCNQVL 420
421 FTCYMASEHS SDETRQCAEG LAKNVNSSHC GIFIDTIVTS ILKVFNVAYG FMPSFQSPDN 480
481 RETMALQNIQ ARIRMVLSYL FAQLALVSHK RPGGLLVLGT ANVDESLVGY LTKYDCSSAD 540
541 INPIGSVSKR DLRQFLEIAY EKYGMAALRC VIDSTPTAEL KPLVDGKVAQ TDEAEIGLTY 600
601 DELSVIGRLR KPGGMGPYGM FLKLLQLWGD KYSIDEIEEK VNKFFWRYRV NRHKATVSTP 660
661 AIHAENYSPD DHRNDHRPFL YPDFSYQFER IREKVVELKK NST |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
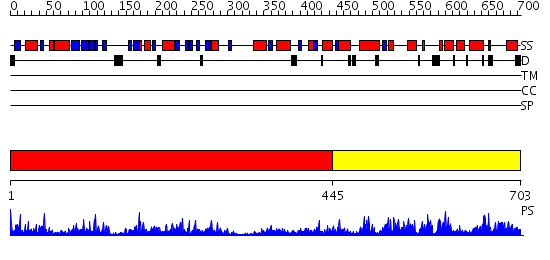
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..444] | 61.39794 | NIT-FHIT fusion protein, C-terminal domain; NIT-FHIT fusion protein, N-terminal domain | |
| 2 | View Details | [445..703] | 61.69897 | NH3-dependent NAD+-synthetase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)